Back to Archived Journals » Advanced Health Care Technologies » Volume 3
Nanomaterial-based biosensors for biological detections
Authors Su H, Li S, Jin Y, Xian ZY, Yang D, Zhou W, Mangaran F, Leung F, Sithamparanathan G, Kerman K
Received 13 September 2016
Accepted for publication 26 July 2017
Published 3 October 2017 Volume 2017:3 Pages 19—29
DOI https://doi.org/10.2147/AHCT.S94025
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Gozde Durmus
Han Su, Shaopei Li, Yiyun Jin, Ziyue Xian, Duowen Yang, Wenxia Zhou, Franklin Mangaran, Felicia Leung, Gobika Sithamparanathan, Kagan Kerman
Department of Physical and Environmental Science, University of Toronto Scarborough, Toronto, ON, Canada
Abstract: Nanomaterial-based biosensors have become one of the major topics in the field of diagnostics. With the growing demand on devices with improved sensitivity and selectivity, rapid response time, and low cost, four categories of nanomaterials have become popular in biosensor research: gold nanoparticles, graphene, carbon nanotubes, and photonic crystals. The ongoing research has brought new designs of biosensors based on nanomaterials, which have greatly improved the potential of field-deployable microfabricated devices. This review describes the recent technologies employing the aforementioned nanomaterials for electrochemical detection of biomolecules, including glucose, DNA, protein, toxins, and so on. We envisage that miniaturized lab-on-a-chip devices employing these nanomaterials will soon be an essential part of our daily life.
Keywords: biosensor, nanomaterial, gold nanoparticle, graphene, carbon nanotube, photonic crystal
Introduction
A biosensor is a device that combines a sensitive biological recognition component and a physical transducer to detect analytes of interest. Analysis results are displayed through transforming the biological reaction into a measurable signal, which can be used for qualitative and quantitative determinations. The biological recognition component part of biosensors usually includes nucleic acids, enzymes, antibodies, receptors, microorganisms, cells, tissues, and even some biomimetic structures. Physical transducers vary significantly with the source of the quantifiable signal, and utilize mostly optical and electrochemical systems. In addition, as demands increase, all constituents in the biosensors can be designed and manufactured in large quantities at a low cost to satisfy the needs of users.1 In 1953, Clark et al2 first published a paper containing the fundamental ideas of a “biosensor”. Moreover, in 1967, Updike and Hicks3 successfully reported a biosensor for glucose detection. From that moment on, with characteristics such as specificity, selectivity, and cost-savings with simple operation, real-time analysis, and continuous use, various types of biosensors were developed rapidly through the second half of century, and have become widely used in related medical, environmental, and forensic fields.4
Nanomaterials range between 1 and 100 nm in size. Nanotechnology has gone through a booming development since 1981.5 After nanotechnology was introduced into biosensors field, sensitivity and other analytical characteristics of biosensors have been greatly enhanced. Among all nanomaterials, gold nanoparticles (AuNPs), carbon nanotubes (CNTs), graphene (GR), and photonic crystals (PCs) stood out due to their unique properties, and are reviewed in this report. The advantages of high stability, superior biocompatibility, high surface energy, strong amplification effect on signals, etc., made them excellent choices for biosensor applications.6–18
For this review, literature search was conducted in PubMed, Web of Science, as well as Google Scholar. Various types of sensors for biological detections are discussed in this review. Four categories of nanomaterials that were searched in detail include “gold nanoparticles”, “carbon nanotubes”, “graphene”, and “photonic crystals”. Publications were reviewed to identify the different methods and modifications of biosensors that employed nanomaterials.
AuNP-based biosensors
One of the most commonly used nanomaterials in the field of biosensors involves colloidal AuNPs. Development of optical and electrochemical biosensors has become very popular due to the unique properties of AuNPs. A major advantage of using AuNPs in biosensors is the increase in detection signal, or response, for analytes that are especially low in concentration. For example, a biosensor based on a boron-doped diamond (BDD) electrode has recently been constructed by Wei et al19 to detect the presence of organophosphate pesticides (OPs), which are harmful for human health, but used widely in agricultural industry. The BDD was modified with carbon spheres (CSs) that were coated with AuNPs. Followed by the addition of acetylcholinesterase (AChE), the detection of model compounds, chlorpyrifos and methyl parathion, was accomplished using differential pulse voltammetry (DPV) by transferring AChE/AuNPs-CSs/BDD that was incubated in the presence of the model compounds, to an electrochemical cell containing 1.0 mM acetylthiocholine chloride as substrate. Oxidation of thiocholine and hydrolysis product of acetylthiocholine chloride resulted in the peak current which indicated the inhibition action of OPs. The involvement of AuNP showed significant improvement by enhancing the detection signal, which could detect OPs as low as 10−13 M. Moreover, Wei and Wang20 also added 1-(4-sulfonic acid) butyl-3-methylimidazolium hydrogen sulfate to the AuNPs-CSs to further increase signal by 1.5-fold.20
As shown in Figure 1, carcinoembryonic antigens (CEAs) were also detected by using two oligonucleotide aptamers.21 Firstly, aptamer-1 (thiol-terminated CEA aptamer 1) was immobilized onto the bare gold electrode. The electrode was then immersed in CEA-containing solution, followed by the addition of 6-ferrocenyl hexanethiol (Fc)-capped AuNPs-aptamer-2 conjugate. Cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), and DPV were applied to the electrode, and the signals were found to be dependent on the concentration of CEA. A concentration of CEA as low as 0.5 ng/mL can be detected, and this sensor showed successful results in human serum.21
  | Figure 1 Schematic diagram of biosensor based on AuNP-based signal amplification for CEA detection using aptamers in sandwich-type assay. Data from Shu et al.21 Abbreviations: AuNP, gold nanoparticle; CEA, carcinoembryonic antigen. |
AuNPs were also conjugated with polyclonal antibodies and added on the polydiacetylene liposomes to enhance fluorescence signals. As shown in Figure 2, Won and Sim22 demonstrated about 100-fold increase in sensitivity by employing AuNPs to detect human immunoglobulin E (hIgE). This sandwich setup allowed detection of hIgE as low as 0.1 ng/mL.
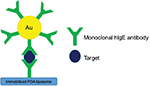  | Figure 2 Schematic diagram of liposome-based biosensor using antibody-conjugated AuNPs for signal amplification. Data from Won and Sim.22 Abbreviations: AuNPs, gold nanoparticles; hIgE, human immunoglobulin E; PDA, polydiacetylene. |
Due to the ever-expanding catalogue of PCR amplification technologies, nucleic acids have been often coupled with AuNPs for detection of disease-related gene sequences. Liu et al23 have tested a method of electroless deposition of AuNPs onto planar gold electrode surface, followed by the immobilization of single-stranded oligonucleotide (probe). The electrode was then immersed into the target DNA-containing solution to allow hybridization to form double-stranded DNA (dsDNA)-modified electrodes. CV was performed to observe the peak current at −0.28 V vs. Ag/AgCl, which corresponded to the assembled dsDNA. By addition of AuNPs, the sensitivity of the electrode could reach 10−11 mol/L as opposed to 10−8 mol/L. The benefit of increased sensitivity was also reported by Hao et al24 in detection of Bacillus anthracis, which is the cause of serious bacterial infection, anthrax,25 using quartz crystal microbalance (QCM). Thiolated DNA probe was immobilized onto the gold surface of QCM via self-assembly. Target DNA with an extension was introduced to the surface and immobilized by hybridization. The signal was amplified with the addition of DNA-conjugated AuNP, which had a complementary sequence with the extension part of target DNA. Hybridization of DNA-conjugated AuNPs with the extension sequence enabled the accumulation of AuNPs on the electrode surface. The amplification of signal by the accumulated AuNPs lowered the detection limit of the bacteria to 3.5 × 102 CFU/mL, as well as provided an alternative assay avoiding the use of carcinogenic reagent ethidium bromide commonly used for gel electrophoresis studies of PCR-amplified nucleic acids.
Another application of AuNP reported by He et al26 involved the immobilization of thiolated single-stranded DNA (ssDNA) and horseradish peroxidase (HRP) on AuNPs (Figure 3). Dual-labeled AuNPs allowed conjugation to the target ssDNA which was also conjugated to the biotinylated capture probe immobilized onto streptavidin-modified surface. As HRP catalyzed the substrate, 3-amino-9-ethylcarbazol, in this case, a red color would appear. The intensity of the red color was correlated with the amount of target DNA in the sample. This colorimetric method detected as low as 0.025 ng/mL human genomic DNA, much lower than most other methods.27
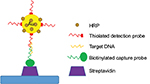  | Figure 3 Schematic illustration of dual-labeled AuNPs with HRP and DNA for the detection of DNA hybridization. Data from He et al.26 Abbreviations: AuNPs, gold nanoparticles; HRP, horseradish peroxidase. |
AuNPs have also been coupled with other nanomaterials such as GR and CNTs for the detection of different analytes. In a study by Azzouzi et al,28 a biosensor was developed for measuring L-lactate using AuNPs anchored on reduced graphene oxide (RGO-AuNPs) and L-lactate dehydrogenase (LDH), over a screen-printed carbon electrode. RGO-AuNPs were prepared by adding RGO to HAuCl4 solution. A mixture of LDH solution and sol–gel matrix was prepared from tetramethoxysilane and methyltrimethoxysilane. RGO-AuNPs suspension was added to the mixture, which was then deposited onto the surface of the working electrode. As LDH enzyme catalyzed the oxidation of lactate in the presence of NAD+, the conversion to pyruvate and NADH was detected amperometrically with 0.13 µM as the detection limit.
In a study by Wang et al,29 the development of a super-sandwich electrochemical DNA biosensor was reported. Glassy carbon electrode (GCE) was immobilized with RGO-AuNPs bound with capture DNA probe with a thiol label. The sandwich structure was formed containing “capture-target-signal probe”. An auxiliary probe was then added to produce long concatamers with methylene blue (MB) as the redox-active intercalator. DNA hybridization was detected using DPV, and the peak current changes of MB were correlated with the logarithm of target DNA concentrations. This biosensor could detect concentrations of DNA as low as 35 aM. Good selectivity was achieved for mismatched target DNA detection.
As one of the most traditional applications of biosensors, glucose detection is very popular in this field. An innovative glucose biosensor, developed by Devasenathipathy et al,30 involved the coupling of biopolymer pectin-stabilized AuNPs electrodeposited on graphene and multiwalled carbon nanotubes (GR-MWCNTs/AuNPs) as shown in Figure 4. The biosensor was prepared by adding multiwalled carbon nanotubes (MWCNTs) in graphite oxide (GO) and ultrasonicating them to obtain GO-MWCNTs. The GO-MWCNTs were then added onto GCE surface, followed by transferring the electrode to an electrochemical cell containing pectin and HAuCl4. After the consecutive CVs at 50 mV/s from +1.40 to −1.40 V, GO was reduced to GR as Au3+ was reduced and electrodeposited on GR-MWCNTs. Glucose oxidase (GOx) was then added to the electrode. Concentration of glucose was measured by CV with a detection limit of 4.1 µM. In practice, the concentration of glucose determined in a blood sample by the GR-MWCNTs/AuNPs-GOx electrode is 5.16 mM as opposed to 4.98 mM that could be detected by an optical biosensor, demonstrating similar results.
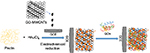  | Figure 4 Schematic flow diagram of the preparation of GR-MWCNTs/AuNPs/GOx film-modified biosensor for the electrochemical detection of glucose. Data from Devasenathipathy et al.30 Abbreviations: GR, graphene; MWCNTs, multiwalled carbon nanotubes; AuNPs, gold nanoparticles; GOx, glucose oxidase; GO, graphene oxide; GCE, glassy carbon electrode. |
GR-based biosensors
GR is made of a single layer of sp2 carbon bond atoms that arrange into a 2D lattice that is known for its electron mobility, thermal conductivity, high surface area, and electrical conductivity. These characteristics of GR allow rapid electron transfer for the detection of biomolecules using biosensors. Problems such as the inconsistent signal amplification of metallic nanoparticles and the metallic impurities in CNTs were not observed in GR-based biosensors.
As mentioned in the previous section, GR has often been coupled with AuNPs in biosensor designs. An elegant method biosensor was prepared by Gupta et al31 using mercaptophenyl boronic acid (MBA)-terminated Ag@AuNPs/graphene oxide (Ag@AuNPs-GO) nanocomposites in connection with surface-enhanced Raman spectroscopy (SERS). AgNO3 was reduced to form silver nanoparticles (AgNPs) followed by the addition of HAuCl4 solution to form Ag@AuNPs. Synthesized GO was activated via 1-ethyl-3-(3-dimethylamino-prophy) carbodiimide hydrochloride (EDC) on carboxylate groups of GO. Ag@AuNPs was then added to the functionalized GO to form a homogeneous mixture through sonication. MBA solution was then added to the Ag@AuNPs-GO nanocomposite, followed by the immobilization of GOx. Detection of glucose was achieved using SERS with a detection limit of 0.33 mM. Liu et al32 also reported a method for glucose biosensor using GO which was biocompatible. In this design, the attachment of GOx to GO was accomplished via EDC and N-hydroxyl succinimide (NHS) activation. Glucose detection was made using chronoamperometric measurements. Biocompatibility was tested by seeding an ARPE-19 cell line derived from human retinal pigment epithelium, onto the GO substrates. This report demonstrated the expanded application of GR as a biocompatible platform for biosensors.
Another coupling of GR and AuNPs was demonstrated by Zhang et al33 in a novel electrochemical biosensor based on AuNPs/thionine-GR nanocomposite to detect thrombin. Thionine-GR was immobilized on a GCE, followed by the addition of AuNPs. Aptamer oligonucleotide was then immobilized onto the surface, which was used to capture the analyte, thrombin. CV, EIS, and DPV were performed for the characterization of electrode surface with a detection limit of 0.093 nM. GR-based biosensors have also been used in detection of hydrogen peroxide. Wang et al34 reported a biosensor using biphenyldimethanethiol (BPT)/AuNPs/GR/HRP composite. Au electrode was immersed in BPT solution, which was then coated with AuNPs/GR composite suspension. HRP was added on the modified electrode surface. CV was employed in the electrocatalysis of HRP, which can detect H2O2 as low as 1.5 μM. As an alternative to enzyme-modified electrodes, Li et al35 developed a hydrogen peroxide sensor by using MnO2/GO nanocomposites. Amperometric response of MnO2/GO electrode enabled the detection of H2O2 concentration as low as 0.8 µM, exceeding the detection limit of traditional enzyme-modified electrodes. Xu et al36 covalently attached GR onto cysteamine-modified Au surface. Then, the GR layer was coupled with α-synuclein (α-syn) protein, which is related to Parkinson’s disease. Serum with specific α-syn antibodies was then applied onto the surface. Autoantibody concentration was determined using EIS with a detection limit of 1.2 pM.
As shown in Figure 5, bacterial DNA detection could also be accomplished using GR-based biosensors. Zainudin et al37 described the drop-casting of RGO suspension onto GCE, followed by addition of 1-pyrenebutyric acid (PyBA). The carboxyl group of PyBA was activated via EDC/NHS, and then modified with probe DNA to detect Escherichia coli cells. CV and EIS were used for monitoring hybridization. The detected DNA concentration was converted into the number of E. coli cells. The biosensor could detect as low as 4.22 × 105 cells/mL, which is comparable to other traditional methods such as ELISA-based ones.
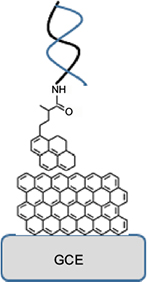  | Figure 5 Schematic illustration of bacterial DNA detection using RGO-modified GCE for detection of DNA hybridization. Data from Zainudin et al.37 Abbreviations: RGO, reduced graphene oxide; GCE, glassy carbon electrode. |
CNT-based biosensors
CNTs comprise carbon atoms arranged hexagonally into six-member carbon rings. These rings interlock and form a GR sheet, which then forms seamless cylindrical tubes known as CNTs.13,14 CNTs can be divided into two categories based on the layering structure of GR sheet: single-walled carbon nanotubes (SWCNTs), which contain only one layer of GR sheet, and MWCNTs, which contain multiple rolled GR layers.13–15
As interest in the development of glucose biosensors never ceases to exist, extensive studies have been conducted to evaluate various CNT-based glucose biosensors. GOx-conjugated CNTs have been utilized frequently. In the classic detection system, GOx generates H2O2 in the presence of glucose. The peroxide species are then further reduced by HRP that is connected to an electrode to produce an electrical signal. In a recent study as described in Figure 6, cytochrome C was immobilized onto MWCNTs followed by GOx and Nafion, respectively, to create an enzyme-modified biosensor with hydroquinone as a redox mediator.38
  | Figure 6 Schematic representation of the construction of GCE/MWCNT-Cyt C/GOx/Naf biosensors for electrochemical detection of glucose. Data from Eguilaz et al.38 Abbreviations: GCE, glassy carbon electrode; MWCNT, multiwalled carbon nanotube; Cyt C, cytochrome C; GOx, glucose oxidase; Naf, Nafion. |
The enzyme-modified biosensor displayed a minimum detection limit of 8 µM glucose. However, the efficiency of glucose detection using a complicated enzymatic system might be hindered by the amount of immobilized protein on the CNTs.39,40 Low amount of immobilized proteins might cause reproducibility issues. To compensate for the low immobilization efficiency, signal amplification can be achieved by further crosslinking additional enzymes on CNTs. As reported by Kwon et al.40 GOx was covalently linked to SWCNTs, and then excessive GOx was further crosslinked onto the covalently attached population using glutaraldehyde as a crosslinker. Additional enhancement of signal amplification was accomplished with the addition of AuNPs. AuNP-CNT-enzyme-modified biosensors had a sensitivity of 24.5 µA/mM.
Though the CNT-enzyme-modified biosensors are highly sensitive, they usually lack stability over time.15 In addition, the design of CNT-enzyme-modified biosensors is laborious.15,41 Therefore, several studies examined the potential of using metal nanoparticles as a substitution of enzyme. As shown in Figure 7, Ensafi et al41 presented a study on a nonenzymatic sensor system using AgNPs. MWCNTs were functionalized and reacted with SOCl2, and then subjected to reflux in ethylenediamine, cyanuric chloride, and diethylenetriamine in nitrogen atmosphere, respectively. AgNO3 was added in the final step with NaBH4 to complete the immobilization of AgNPs.41
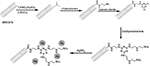  | Figure 7 Schematic flowchart of the construction of AgNP-modified MWCNTs for electrochemical detection of glucose. Data from Ensafi et al.41 Abbreviations: AgNP, silver nanoparticle; MWCNTs, multiwalled carbon nanotubes. |
AgNP-based glucose biosensors had a detection limit of 0.03 µM. In addition, AgNP-MWCNT-based biosensor showed comparable results to an automated glucose analyzer, when it was subjected to the detection of glucose level in human blood samples. Furthermore, the AgNP-MWCNT-based biosensor demonstrated a negative response under the optimized conditions, when other sugar molecules such as sucrose and fructose were present in the sample. However, AgNP-MWCNT biosensor system still required multistep synthesis,41 but Sun et al39 have recently reported a one-step synthesis using ionic liquids as a method for rapid synthesis of nonenzymatic glucose biosensors. MWCNTs were initially functionalized using concentrate nitric acid and then combined with a mixture comprising ethylene and NiCl2. The nickel-containing mixture was deposited onto the GCE surface.39 This system showed a detection limit of 0.89 µM. In addition, after 30 days of fabrication, the current response dropped only about 5%.39 Though the biosensor still lacked high specificity, nonenzymatic CNT-based electrochemical systems proved to be a promising direction for future glucose sensor development.
Yang and Zhang42 utilized SWCNTs in conjunction with MB-modified ssDNA probes as the label, which successfully avoided the interference from the complicated chemical modifications and prevented the denaturation of ssDNA probes. Results showed that, in the presence of 16-base complementary target DNA, MB molecules were removed from the electrode surface and gave a low current response using square wave voltammetry, while giving a high current signal in the absence of DNA. This SWCNT-based biosensor demonstrated the detection of nucleic acids with improved sensitivity comparing to biosensors without SWCNT modifications, a strong sequence specificity toward even single-base-mismatched DNA, and a large linear dynamic range with a detection limit as low as 1.0 pM.
A label-free and signal-off electrochemical DNA-wrapped MWCNT biosensor coupled with hairpin oligonucleotide switch was constructed by Shahrokhian et al43 to detect specific DNA sequences using [Fe(CN)6]3−/4− as a redox probe. CV and DPV showed an increase in peak separation and decrease in faradic peak current when negatively charged DNA was immobilized onto the electrodes. The enhanced response was due to the π–π interaction between ssDNA and MWCNT and increased surface conductivity. DPV peak current showed that the developed biosensor provided sensitive and stable electrochemical responses with a good linear correlation range from 0.1 mM to 10 pM for the target DNA. The biosensor also showed high specificity for the discrimination of a noncomplementary DNA sequence.
In another study, Qiu et al44 introduced a CNT-based lateral flow biosensor (LFB) for the detection of DNA fragments. Amine-modified DNA probe was immobilized on the shortened MWCNTs surface via diimide-activated amidation between the carboxylic acid groups on CNTs and amino groups on target DNA. Sandwich-type hybridization was performed on LFB plate, where the captured MWCNTs on the test zone (strep-biotinylated capture probe) and control zone (strep-biotinylated control probe) produced black bands to enable visual detection of DNA sequence. According to the intensity of test line, the CNT-based LFB sensor was capable of detecting 40 pM DNA after systematic optimization studies.
As shown in Figure 8, target DNA was applied on the “sample pad” and then was flown through the “conjugate pad” with MWCNTs-DNA. The first hybridization reaction occurred, forming an MWCNT-DNA-target DNA complex. As the complex further migrated, it was captured and hybridized again with the biotinylated capture DNA probe on the “test line”. Extra MWCNT-DNA conjugates were captured by the biotinylated-control probe. MWCNT-DNA conjugates that were captured gave dark black bands for easy visualization of the test results by naked eye.
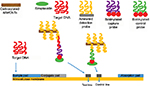  | Figure 8 Schematic representation of MWCNT-based LFB for the detection of DNA hybridization. Data from Qiu et al.44 Abbreviations: MWCNT, multiwalled carbon nanotube; LFB, lateral flow biosensor. |
CNT-based biosensors were also used to trap and detect whole cells. For example, Abdolahad et al45 developed a vertically aligned CNT-based electrical cell impedance biosensor (CNT-ECIS) fabricated with an Si/SiO2 layer, where Ni was deposited as a catalyst for the detection of cancer cells. CNT arrays acted as both adhesive and conductive layer, so that the direct attachment of cell membrane to electrodes led to the enhanced electrical interaction. Also, the piercing of CNT tips to the cells avoided any gap between the cells and conductive MWCNT arrays. Impedance of SW48 cells from grade-IV human colon tumors was measured. Results showed that CNT-ECIS was able to detect cancer cells at a concentration as low as 4000 cells/cm2 and achieved a sensitivity of 1.7 × 10−3 Ohm/cm2 in 30 s. Higher population of attached cells resulted in further changes in CNT-ECIS impedance. Cells undergoing apoptosis could also be monitored using CNT-ECIS sensors.45
Another focus of recent literature on nanomaterial-based biosensors has been the detection of microorganisms. Thuy et al46 introduced an MWCNT dispersion method to detect E. coli O157:H7 using a DNA biosensor. DNA sequences related to E. coli O157:H7 could be detected at a concentration as low as 1 nM with sensitivity around 0.19 nM/mV. García-Aljaro et al47 reported a CNT-based chemiresistive biosensor for detection of bacteria and viruses using parallel-aligned SWNTs bridging two gold electrodes for the detection of E. coli O157:H7 and bacteria T7. SWNTs were functionalized with specific antibodies by covalent immobilization to the non-covalently bound 1-pyrene butanoic acid succinimidyl ester (PBASE). As shown in Figure 9, PBASE was adsorbed onto the SWCNTs bridging two gold electrodes. Specific antibody (e.g. monoclonal anti-E. coli O157: H7 antibody) was covalently bound to PBASE. Linear sweep voltammetry was performed when target microorganism bound to the antibody. E. coli O157:H7 displayed significant increase in resistance with a detection limit of 105 CFU for whole cells and 103 CFU for lysates. Bacteria T7 could also be detected with a limit of 103 PFU/mL. It was concluded that the performance of this biosensor was better for virus detection compared to bacteria due to the smaller size and better diffusion characteristics of viruses.
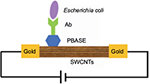  | Figure 9 Schematic representation of CNT-based chemiresistive biosensor for the detection of bacteria and viruses. Data from Thuy et al.46 Abbreviations: CNT, carbon nanotube; Ab, antibody; PBASE, 1-pyrene butanoic acid succinimidyl ester; SWCNTs, single-walled carbon nanotubes. |
PC-based biosensors
PCs are nanostructures that possess a periodic dielectric surface with a photonic band gap, which does not allow light propagation of a specific wavelength range.48,49 Such control and manipulation of light allows optical detection applications to be studied experimentally according to Bragg’s Law.50 These materials are often made from silica or indium oxide, carefully constructed and modified for their chemical and optical properties.51–57 The optical properties of PCs are related to their periodicity, size, and average refractive index.51
Sharma and Sharan52,53 detected high glucose concentration in urine, known as glycosuria, using a PC-based structure with line defects and 2D air holes in a silicon slab. An electromagnetic equation propagation simulation tool was used to model the sensor structure. The difference in decibels in normal urine and glycosuric urine indicated that sensor could be used for the detection of glycosuria. Presence of high glucose concentration may lead to cardiovascular diseases and diabetes. Variation in refractive index of urine in the presence of glucose was detected by passing light through the PC-based biosensor. Results showed the variation in the input refractive index values with different concentrations of glucose in urine.
PC-based biosensors could be used for early diagnosis of viral diseases. Endo et al54 employed a 2D PC-based biosensor for the reflectometric detection of the influenza virus in human saliva. Nanoimprinting technology was used in the fabrication of this PC-based biosensor due to its low cost and high resolution. H1N1 hemagglutinin was utilized for the detection of the influenza virus. As shown in Figure 10, a UV–vis spectrophotometer was utilized to evaluate the optical characteristics of the 2D PC-based biosensor. Changes in reflection intensity were detected upon formation of the antigen–antibody complex.
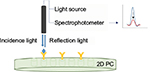  | Figure 10 Illustration of reflectometric detection of influenza virus using antibody-immobilized 2D PC. Data from Endo et al.54 Abbreviation: PC, Photonic crystal. |
A similar optical setup was employed by Cheng et al55 for the detection of DNA hybridization related to a specific point mutation in apolipoprotein E gene using localized surface plasmon resonance (LSPR) and EIS. AuNPs were electrodeposited onto indium tin oxide (ITO) surface using CV. The probe DNA was immobilized onto the AuNPs allowing hybridization with the target DNA. LSPR resulted in a decrease in absorbance when the setup was exposed to mismatch and noncomplementary DNA sequences instead of target (complementary) DNA. EIS also showed a decrease in charge transfer resistance (RCT). Cheng et al56 also conducted a study using ITO by depositing the surface with Au using an electron beam evaporator, forming Au nanopyramids, as shown in Figure 11. The protein (α-syn) was immobilized onto the Au nanopyramids via a linker 3,3′-dithiobios (sulfosuccinimidyl-propionate). Small molecules were screened using electrochemistry and LSPR for their interaction with the immobilized α-syn.
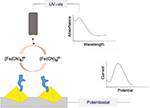  | Figure 11 Schematic representation of electrochemical and LSPR-based detection of α-synuclein aggregation using Au nanopyramid-modified ITO surface. Data from Cheng et al.55 Abbreviations: LSPR, localized surface plasmon resonance; ITO, indium tin oxide; UV-vis, ultraviolet visible. |
Shen et al57 employed the colloidal PCs in a fluoroimmunoassay (Figure 12). It was observed that the colloidal PCs made an excellent substrate, which greatly enhanced sensitivity and accuracy in terms of protein detection. The target avidin was sandwiched between biotin-modified colloidal PCs and the fluorescent-labeled ligand (Atto 425-Biotin). The signal was greatly enhanced by the large surface area of colloidal PCs compared to planar PCs.
  | Figure 12 Schematic illustration of enhanced avidin fluoroimmunoassay on colloidal PCs. Data from Shen et al.57 Abbreviations: PCs, photonic crystals; BSA, bovine serum albumin. |
Detection of myoglobin was also accomplished using PC in a total-internal-reflection configuration (TIR).58 Presence of myoglobin could indicate possible diagnosis of acute myocardial infarction (AMI). The sensor was made from a BK7 glass substrate with five alternating layers of titania and silica and a cavity layer at the surface. The sensor surface was modified to possess amine groups, which were covalently attached with cardiac myoglobin antibodies via EDC/NHS activation. Myoglobin was detected with a detection limit of 70 ng/mL using this method, which fell in the clinical diagnostic level of AMI patients. The PC-TIR system was also used by Guo et al59 in detection of biotinylated analytes through streptavidin-modified silica surfaces. The response between biotinylated analytes and streptavidin was related to the size, amount of streptavidin, and number of free sites on the surface. Molecules with a molecular weight as low as 244 Da could be detected using this biosensor.
A fast, mobile lab-on-a-chip construct was prepared by Han et al60 using PCs to detect staphylococcal enterotoxin B (SEB), one of the most common bacterial toxins associated with food-borne illnesses. ITO-coated glass surfaces were modified with anti-SEB monoclonal antibodies for fluorescence measurements. The procedure required very low amount of sample (10 µL) with short incubation time and was able to detect as low as 1 ng/mL.
Conclusion
Nanomaterial-based biosensors have become more widely available and are increasingly utilized in modern detection technologies. The traditional drawbacks of biosensors such as sensitivity and selectivity have been overcome with the modification of biorecognition layers using nanomaterials. In general, the ongoing research efforts on nanomaterial-based biosensors are recently focusing on reducing the cost and analysis time, while improving sensitivity and selectivity. As these research efforts grow exponentially, it is inevitable that the face of traditional detection technologies in the biomedical field will be drastically altered in the near future. Portable and affordable devices with embedded nanomaterials will soon be available in the market.
Disclosure
The authors report no conflicts of interest in this work.
References
Mavrogiannis N, Crivellari F, Gagnon ZR. Label-free biomolecular detection at electrically displaced liquid interfaces using interfacial electrokinetic transduction (IET). Biosens Bioelectron. 2016;77:790–798. | ||
Clark LC Jr, Wolf R, Granger D, Taylor Z. Continuous recording of blood oxygen tensions by polarography. J Appl Physiol. 1953;6(3):189–193. | ||
Updike SJ, Hicks GP. The enzyme electrode. Nature. 1967;214(5092):986–988. | ||
Wang J. Electrochemical Glucose Biosensors. Chem Rev. 2008;108(2):814–825. | ||
Taylor R, Sylvain C, Todd O, et al. Small particle, big impacts: a review of the diverse applications of nanofluids. J Appl Phys. 2013;113(1):011301. | ||
Fevero G, Fusco G, Mazzei F, Tasca F, Antiochia R. Electrochemical characterization of graphene and MWCNT screen-printed electrodes modified with AuNP for laccase biosensor development. Nanomaterials (Basel). 2015;5(4):1995–2006. | ||
Adams NM, Jackson SR, Haselton FR, Wright DW. Design synthesis and characterization of nucleic-acid-functionalized gold surfaces for biomarker detection. Langmuir. 2012;28:1068–1082. | ||
Sperling RA, Gil PR, Zhang F, Zanella M, Parak WJ. Biological applications of gold nanoparticles. Chem Soc Rev. 2008;37(9):1896–1908. | ||
Kuila T, Bose S, Khanra P, Misha AK, Kim NH, Lee JK. Recent advances in graphene-based biosensors. Biosens Bioelectron. 2011;26(12):4637–4648. | ||
Akbari E, Buntat Z, Kiani MJ, Enzevaee A, Khaledian M. Analytical model of graphene-based biosensors for bacteria detection. Int J Environ Anal Chem. 2015;95(9):847–854. | ||
Radhapyari K, Kotoky P, Das MR, Khan R. Graphene-polyaniline nanocomposite based biosensor for detection of antimalarial drug artesunate in pharmaceutical formation and biological fluids. Talanta. 2013;111:47–53. | ||
Singh C, Ali MA, Sumana G. Green synthesis of graphene based biomaterial using fenugreek seeds for lipid detection. ACS Sustain Chem Eng. 2016;4(3):871–880. | ||
Lin Y, Taylor S, Li H, et al. Advances toward bioapplications of carbon nanotubes. J Mater Chem. 2004;14(4):527–541. | ||
Kalbacova M, Kalbac M, Dunsch L, Kataura H, Hempel U. The study of the interaction of human mesenchymal stem cells and monocytes/macrophages with single-walled carbon nanotube films. Phys Stat Sol B. 2006;243(13):3415–3418. | ||
Yang N, Chen X, Ren T, Zhang P, Yang D. Carbon nanotube based biosensors. Sens Actuators B. 2015;207:690–715. | ||
Aki S, Endo T, Sueyoshi K, Hisamoto H. Plasticized poly(vinyl chloride)-based photonic crystal for ion sensing. Anal Chem. 2014;84(24):11986–11991. | ||
Endo T, Ikeda R, Yanaqida Y, Hatsuzawa T. Stimuli-responsive hydrogen-silver nanoparticles composite for development of localized surface plasmon resonance-based optical biosensor. Anal Chim Acta. 2008;611(2):205–211. | ||
Ma Y, Di J, Yan X, Zhao M, Lu Z, Tu Y. Direct electrodeposition of gold nanoparticles on indium tin oxide surface and its application. Biosens Bioelectron. 2009;24(5):1480–1483. | ||
Wei M, Zeng G, Lu Q. Determination of organophosphate pesticides using an acetylcholinesterase-based biosensor based on a boron-doped diamond electrode modified with gold nanoparticles and carbon spheres. Microchim Acta. 2014;181(1–2):121–127. | ||
Wei M, Wang J. A novel acetylcholinesterase biosensor based on ionic liquid-AuNPs-porus carbon composite matrix for detection of organophosphate pesticides. Sens Actuators B. 2015;211:290–296. | ||
Shu H, Wen W, Xiong H, Zhang X, Wang S. Novel electrochemical aptamer biosensor based on gold nanoparticles signal amplification for the detection of carcinoembryonic antigen. Electrochem Commun. 2013;37:15–19. | ||
Won SH, Sim SJ. Signal enhancement of a micro-arrayed polydiacetylene (PDA) biosensor using gold nanoparticles. Analyst. 2012;137(5):1241–1246. | ||
Liu S, Liu J, Wang L, Zhao F. Development of electrochemical DNA biosensor based on gold nanoparticle modified electrode by electroless deposition. Bioelectrochemistry. 2010;79(1):37–42. | ||
Hao RZ, Song HB, Zuo GM, et al. DNA probe functionalized QCM biosensor based on gold nanoparticle amplification for Bacillus anthracis detection. Biosens Bioelectron. 2011;26(8):3398–3404. | ||
Edwards KA, Clancy HA, Baeumner AJ. Bacillus anthracis: toxicology epidemiology and current rapid-detection methods. Anal Bioanal Chem. 2006;384(1):73–84. | ||
He Y, Zhang S, Zhang X, et al. Ultrasensitive nucleic acid biosensor based on enzyme-gold nanoparticle dual label and lateral flow strip biosensor. Biosens Bioelectron. 2011;26(5):2018–2024. | ||
Mao X, Ma Y, Zhang A, Zhang L, Zeng L, Liu G. Disposable nucleic acid biosensors based on gold nanoparticle probes and lateral flow strip. Anal Chem. 2009;81(4):1660–1668. | ||
Azzouzi S, Rotariu L, Benito AM, Maser WK, Ali MB, Bala C. A novel amperometric biosensor based on gold nanoparticle anchored on reduced graphene oxide for sensitive detection of L-lactate tumor biomarker. Biosens Bioelectron. 2015;69:280–286. | ||
Wang J, Shi A, Fang X, Han X, Zhang Y. An ultrasensitive supersandwich electrochemical DNA biosensor based on gold nanoparticles decorated reduced graphene oxide. Anal Biochem. 2015;469:71–75. | ||
Devasenathipathy R, Mani V, Chen SM, et al. Glucose biosensor based on glucose oxidase immobilized at gold nanoparticles decorated graphene-carbon nanotubes. Enzyme Microb Technol. 2015;78:40–45. | ||
Gupta VK, Atar N, Yola ML, et al. A novel glucose biosensor platform based on Ag@AuNPs modified graphene oxide nanocomposite and SERS application. J Colloid Interface Sci. 2013;406:231–237. | ||
Liu Y, Yu D, Zeng C, Miao Z, Dai L. Biocompatible graphene oxide based glucose biosensors. Langmuir. 2010;26(9):6158–6160. | ||
Zhang Z, Luo L, Zhu L, Ding Y, Deng D, Wang Z. Aptamer-linked biosensor for thrombin based on AuNPs/thionine-graphene nanocomposite. Analyst. 2013;138(18):5365–5370. | ||
Wang T, Zhu Y, Li G, et al. A novel hydrogen peroxide biosensor based on the BPT/AuNPs/graphene/HRP composite. Sci China Chem. 2011;54(10):1645–1650. | ||
Li L, Du Z, Liu S, et al. A novel nonenzymatic hydrogen peroxide sensor based on MnO2/graphene oxide nanocomposite. Talanta. 2010;82(5):1637–1641. | ||
Xu Q, Cheng H, Lehr J, Patil AV, Davis JJ. Graphene oxide interfaces in serum based autoantibody quantification. Anal Chem. 2015;87(1):346–350. | ||
Zainudin N, Hairul ARM, Yusoff MM, Tan LL, Chong KF. Impedimetric graphene-based biosensor for the detection of Escherichia coli DNA. Anal Methods. 2014;6(19):7935–7941. | ||
Eguílaz M, Venegas CJ, Gutiérrez A, Rivas GA, Bollo S. Carbon nanotubes non-covalently functionalized with cytochrome c: a new bioanalytical platform for building bienzymatic biosensors. Microchem J. 2016;128:161–165. | ||
Sun A, Zheng J, Sheng Q. A highly sensitive non-enzymatic glucose sensor based on nickel and multi-walled carbon nanotubes nanohybrid films fabricated by on-step co-electrodeposition in ionic liquids. Electrochim Acta. 2012;65:64–69. | ||
Kwon KY, Yang SB, Kong BS, Kim J, Jung HT. High-performance biosensors based on enzyme precipitate coating in gold nanoparticle-conjugated single-walled carbon nanotube network films. Carbon. 2010;48(15):4504–4509. | ||
Ensafi AA, Zandi-Atashbar N, Rezaei B, Chermahini ME, Moshiri P. Non-enzymatic glucose electrochemical sensor based on silver nanoparticle decorated organic functionalized multiwall carbon nanotubes. RSC Adv. 2016;6(65):60926–60932. | ||
Yang K, Zhang CY. Simple detection of nucleic acids with a single-walled carbon-nanotube-based electrochemical biosensor. Biosens Bioelectron. 2011;28(1):257–262. | ||
Shahrokhian S, Salimian R, Kalhor HR. A simple label-free electrochemical DNA biosensor based on carbon nanotube–DNA interaction. RSC Adv. 2016;6(19):15592–15598. | ||
Qiu W, Xu H, Takalkar S, et al. Carbon nanotube-based lateral flow biosensor for sensitive and rapid detection of DNA sequence. Biosens Bioelectron. 2015;64:367–372. | ||
Abdolahad M, Taghinejad M, Taghinejad H, Janmaleki M, Mohajerzadeh SA. Vertically aligned carbon nanotube-based impedance sensing biosensor for rapid and high sensitive detection of cancer cells. Lab Chip. 2012;12(6):1183–1190. | ||
Thuy NT, Tam PD, Tuan MA, et al. Detection of pathogenic microorganisms using biosensor based on multi-walled carbon nanotubes dispersed in DNA solution. Curr Appl Phys. 2013;12(6):1553–1560. | ||
García-Aljaro C, Cella LN, Shirale DJ, et al. Carbon nanotubes-based chemiresistive biosensors for detection of microorganisms. Biosens Bioelectron. 2010;26(4):1437–1441. | ||
John S. Strong localization of photons in certain disordered dielectric superlattices. Phys Rev Lett. 1987;58(23):2486–2489. | ||
Yablonovitch E. Photonic band-gap crystals. J Phys Condens Matter. 1993;5:2443–2460. | ||
Vlasov YA, O’Boyle M, Hamann HF, McNab SJ. Active control of slow light on a chip with photonic crystal waveguides. Nature. 2005;438(7064):65–69. | ||
Xu X, Asher SA. Synthesis and utilization of monodisperse hollow polymeric particles in photonic crystals. J Am Chem Soc. 2004;126(25):7940–7945. | ||
Sharma P, Sharan P. An analysis and design of photonic crystal based biochip for detection of glycosuria. IEEE Sens J. 2015;15(10):5569–5575. | ||
Sharma P, Sharan P. Design of photonic crystal-based biosensor for detection of glucose concentration in urine. IEEE Sens J. 2015;15(2):1035–1042. | ||
Endo T, Ozawa S, Okuda N, Yanagida Y, Tanaka S, Hatsuzawa T. Reflectometric detection of influenza virus in human saliva using nanoimprint lithography-based flexible two-dimensional photonic crystal biosensor. Sens Actuators B. 2010;148:269–276. | ||
Cheng XR, Hau BY, Endo T, Kerman K. Au nanoparticle-modified DNA sensor based on simultaneous electrochemical impedance spectroscopy and localized surface plasmon resonance. Biosens Bioelectron. 2014;53:513–518. | ||
Cheng XR, Wallace GQ, Lagugne-Labarthet F, Kerman K. Au nanostructured surfaces for electrochemical and localized surface plasmon resonance-based monitoring of α-synuclein-small molecule interactions. ACS Appl Mater Interfaces. 2015;7:4081–4088. | ||
Shen W, Li M, Xu L, et al. Highly effective protein detection for avidin-biotin system based on colloidal photonic crystals enhanced fluoroimmunoassay. Biosens Bioelectron. 2011;26(5):2165–2170. | ||
Zhang B, Tamez-Vela JM, Solis S, et al. Detection of myoglobin with an open-cavity-based label-free photonic crystal biosensor. J Biomed Eng. 2013;2013:808056. | ||
Guo Y, Ye JY, Divin C, et al. Real-time biomolecular binding detection using a sensitive photonic crystal biosensor. Anal Chem. 2010;82(12):5211–5218. | ||
Han JH, Kim HJ, Sudheendra L, Gee SJ, Hammock BD, Kennedy IM. Photonic crystal lab-on-a-chip for detecting staphylococcal enterotoxin B at low attomolar concentration. Anal Chem. 2013;85(6):3104–3109. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
