Back to Journals » International Journal of Nanomedicine » Volume 14
Long noncoding RNA expression analysis reveals the regulatory effects of nitinol-based nanotubular coatings on human coronary artery endothelial cells
Authors Shang P, Chen G , Zu G, Song X, Jiao P, You G, Zhao J, Li H , Zhou H
Received 3 February 2019
Accepted for publication 4 April 2019
Published 7 May 2019 Volume 2019:14 Pages 3297—3309
DOI https://doi.org/10.2147/IJN.S204067
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Pan Shang,1,* Gan Chen,1,* Guannan Zu,2 Xiang Song,1 Peng Jiao,2 Guoxing You,1 Jingxiang Zhao,1 Hongyi Li,2 Hong Zhou1
1Academy of Military Medical Sciences, Institute of Health Service and Transfusion Medicine, Beijing 100850, People’s Republic of China; 2Photoelectrochemical Research Group, School of Materials Science and Engineering, Beijing University of Technology, Beijing 100124, People’s Republic of China
*These authors contributed equally to this work
Background: Cardiovascular disease (CVD) is the leading cause of mortality all over the world. Vascular stents are used to ameliorate vascular stenosis and recover vascular function. The application of nanotubular coatings has been confirmed to promote endothelial cell (EC) proliferation and function. However, the regulatory mechanisms involved in cellular responses to the nanotubular topography have not been defined. In the present study, a microarray analysis was performed to explore the expression patterns of long noncoding RNAs (lncRNAs) in human coronary artery endothelial cells (HCAECs) that were differentially expressed in response to nitinol-based nanotubular coatings.
Materials and methods: First, anodization was performed to synthesize nitinol-based nanotubular coatings. Then, HCAECs were cultured on the samples for 24 h to evaluate cell cytoskeleton organization. Next, total RNA was extracted and synthesized into cRNA, which was hybridized onto the microarray. GO analysis and KEGG pathway analysis were performed to investigate the roles of differentially expressed messenger RNAs (mRNAs). Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) was performed to validate the expression of randomly selected lncRNAs. Coexpression networks were created to identify the interactions among lncRNAs and the protein-coding genes involved in nanotubular topography-induced biological and molecular pathways. Independent Student’s t-test was applied for comparisons between two groups with statistical significance set at p<0.05.
Results: 1085 lncRNAs and 227 mRNAs were significantly differentially expressed in the nitinol-based nanotubular coating group. Bioinformatics analysis revealed that extracellular matrix receptor interactions and cell adhesion molecules play critical roles in the sensing of nitinol-based nanotubular coatings by HCAECs. The TATA-binding protein (TBP) and TBP-associated transfactor 1 (TAF1) are important molecules in EC responses to substrate topography.
Conclusion: This study suggests that nanotubular substrate topography regulates ECs by differentially expressed lncRNAs involved extracellular matrix receptor interactions and cell adhesion molecules.
Keywords: nitinol titanium dioxide nanotubes, RNA sequencing, long noncoding RNA, nanotopography, molecular networks
Introduction
Cardiovascular disease is the leading cause of death all over the world.1 Various cardiovascular prostheses are often employed to treat and reconstruct damaged cardiovascular structures, such as cardiovascular vessels and heart tissues. However, clinical complications of the prostheses, such as in-stent restenosis and late stent thrombosis, frequently occur after implantation. Although numerous surface modification strategies have been developed, preventing these complications remains challenging.
Previous studies have demonstrated that rapid endothelialization on the surfaces of vascular prostheses is the most effective way to inhibit the above complications.2 There are many factors regulating rapid endothelialization on the surface of a prosthesis.3 One of the most significant factors is the interaction between the biomaterial surface and the ECs. It has been reported that the rate and quality of endothelialization on vascular prostheses depend on these interactions.4 Many factors regarding cell-substrate interactions are known to affect endothelial cellular adhesion, proliferation, spreading and other processes. Surface topography is a critical parameter among these factors.5 Numerous vascular stents with various topographic features have been designed to study the interactions between ECs and the cell-substrate interface.6 One of the most promising designs involves an upright nanotubular coating on the surface of a metallic substrate created via a simple anodization process.7 Several studies have investigated the behaviors and functions of ECs in response to nanotubular topography with titanium nanotubes.7–9 Nitinol is the universal clinical vascular stent material because of its superelasticity and blood compatibility; however, there have been few studies on nitinol nanotubes, although one recent study has shown that nitinol-based nanotubular coatings can improve endothelialization by promoting cell migration and the spreading of human aortic endothelial cells.10 Nevertheless, the mechanisms regulating the expression of genes in ECs responding to nitinol-based nanotubular coatings have not been defined.
Recent studies have shown that lncRNAs are functional RNA molecules longer than 200 nucleotides that are generally not translated into proteins but can regulate the expression and function of protein-coding genes by different mechanisms.11 Many lncRNAs have been implicated in cardiovascular pathophysiology and have been inferred as potential therapeutic targets.12 Recent evidence indicates that lncRNAs modulate the proliferation and migration phenotype of ECs, and aberrant expression of lncRNAs is closely associated with EC function13 and death.14 Therefore, it is significant to determine the roles of lncRNAs in promoting EC proliferation and function in response to nitinol-based nanotubular coatings.
In previous studies, we investigated the effect of TiO2 nanotubes on the osteogenic differentiation process and the biological attachment mechanism of bone implants.15,16 On this basis, we are interested in the roles of lncRNAs in EC responses to nanotubular topography. Therefore, we cultured HCAECs on nitinol-based nanotubular coatings and utilized RNA-sequencing technology to analyze the expression patterns of lncRNAs and mRNAs in the HCAECs. Microarrays revealed that the nitinol nanotubes enhanced HCAEC motility via the sensing of nanotopographical cues by molecules involved in extracellular matrix receptor interaction and cell adhesion. Networks generated from significantly differentially expressed lncRNAs indicated that TBP and TAF1 are critical molecules in the interaction of ECs with nitinol-based nanotubular topography. Our results confirmed that 1085 differentially expressed lncRNAs participate in the regulation of HCAECs induced by nitinol-based nanotubular coatings.
Material and methods
Synthesis and characterization of nitinol-based nanotubular coatings
Nitinol foils were purchased (58.72 wt% Ni, superelastic, pickled surface, PEIER, Jiangyin, China) and were cut into 2×2-cm pieces. Then the nitinol foils were cleaned by ultrasonication in acetone and ethanol (Sinopharm Group Co., Ltd, Beijing, China). The foils were dried under nitrogen, and the control nitinol substrates and nanotubular substrates were labeled CK and NT, respectively. Anodization was conducted at 20 °C in a Teflon container. The nitinol foil is the working anode while graphite foil is the counter electrode. Anodization was performed as described in our previous study on titanium nanotubes,17 with slight modifications. Briefly, the electrolyte contained 2 g of NH4F (Sinopharm Group Co., Ltd), 1000 mL of ethylene glycol (Sinopharm Group Co., Ltd) and 10 mL of Millipore water. The anodization voltage was 30 V and the duration was 30 min. Nanotubular substrates were rinsed in ethanol and Millipore water after anodization. Samples were subsequently annealed in a muffle roaster (CINITE, Beijing, China) for 2 h at a temperature of 500 °C to crystallize the nanotubes. Each substrate was sterilized with 75% ethanol and UV radiation before cell culture. The nanotubular coatings were observed with a scanning electron microscope (JEOL-JSM7001F, Tokyo, Japan). In addition, surface hydrophilicity was evaluated by measuring water contact angles (DataPhysics OCA15Pro, Filderstadt, Germany). Because nanotubular arrays are directly grown on nitnol sheets with 0.3mm thickness and opaque, which is far more than the 100nm thickness required for transmission electron microscope (TEM) study. Therefore, the nanotubular arrays on nitinol sheets can not support TEM study. The diagram is shown in
Cellular experiments
HCAECs were purchased (ScienCell, San Diego, California, USA) and cultured under the manufacturer’s instructions. Cells in passages 2–5 were used. The cell culture medium for the HCAECs was Endothelial Cell Medium (ScienCell) supplemented with endothelial cell growth factor, 20% FBS and 100 U/mL penicillin/100 µg/mL streptomycin. Cells were cultured in an incubator containing 5% (v/v) CO2 at 37 °C and culture medium was renewed every 2 days. For the extraction of total RNA, HCAECs were seeded onto the substrates, without protein pre-coating, and were then cultured for 48 h.
To evaluate cell cytoskeleton organization, HCAECs were cultured on the samples for 24 h based on previous article7 and were then fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 15 min. Then the cells were permeabilized using 0.1% Triton X-100 (Sigma-Aldrich) in PBS containing 1% bovine serum albumin (Sigma-Aldrich) for 10 min. Subsequently, the cells were labeled for F-actin using phalloidin conjugated to tetramethyl rhodamine isothiocyanate (Sigma-Aldrich) for 20 min. Also, the cell nuclei were stained with 4–6-diamidino-2-phenylindole (Invitrogen, Carlsbad, USA) for 15 min. After staining, the samples were washed three times in PBS. Finally, the samples were mounted on microscope slides for observation with a confocal laser scanning microscopy (Nikon A1, Tokyo, Japan).
To confirm the difference in cell phenotype between the CK and NT groups, immunofluorescent analyses of CD31, vWF, and vinculin were conducted. HCAECs were cultured on the samples for 72 h, and then fixed with 4% paraformaldehyde (Sigma-Aldrich) in PBS for 15 min. Then, the cells were permeabilized using 0.1% Triton X-100 (Sigma-Aldrich) in PBS containing 1% bovine serum albumin (Sigma-Aldrich) for 10 min. Next, the cells were incubated with CD31 mAb (Cell Signaling Technology, Danvers, MA, USA), vWF polyclonal antibody (Proteintech, Chicago, USA), and vinculin mAb (Proteintech) at 4 °C overnight. Subsequently, the cells were stained with Alexa Fluor 488/555-conjugated secondary antibody (Cell Signaling Technology). The cell nuclei were also stained with 4′,6-diamidino-2-phenylindole (Invitrogen) for 15 min. Following each step, the cells were washed three times with PBS. After that, the expression of adhesion molecules was determined by confocal laser scanning microscopy (Nikon A1).
For quantification of cytoskeletal features, ImageJ software was used to quantify cytoskeletal elongation. Elongation was quantified by measuring the long and short axes of the cell and calculating the aspect ratio (long to short axis ratio). Ten cells in each group were randomly selected for statistical analysis.
Microarray analysis
Total RNA was extracted from each sample in TRIzol Reagent (TaKaRa, Tokyo, Japan) with an Ultrapure RNA Kit (CWBIO, Beijing, China) under the manufacturer’s instructions. Total RNA was quantified with a NanoDrop ND-2000 (Thermo Scientific, Waltham, MA, USA). Briefly, total RNA was synthesized into double-stranded cDNA. Then the cDNA was synthesized into cRNA labeled with Cyanine-3-CTP, which was hybridized onto the microarray. After washing, the arrays were scanned with an Agilent G2505C Microarray Scanner (Agilent Technologies, Palo Alto, USA).
The array images were analyzed with Feature Extraction software (version 10.7.1.1, Agilent Technologies). The raw data were normalized via quantile algorithm and analyzed with the Agilent GeneSpring (version 14.8, Agilent Technologies). The probes that had at least 1 condition out of 2 conditions flagged as “p” were chosen for further analysis. Then differentially expressed genes and lncRNAs were identified by fold changes and p-values calculated with t-tests. The threshold for upregulated and downregulated genes was a fold change ≥2.0 and a p-value ≤0.05. Finally, hierarchical clustering was conducted to display the expression patterns of the distinguishable genes among the samples.
Quantitative real-time PCR validation
qRT-PCR was carried out with a Bio-Rad thermocycler and SYBR Green Real-Time PCR Master Mix (Toyobo, Tokyo, Japan) under the manufacturers’ instructions. The reaction conditions were as follows: incubation at 95 °C for 1 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 15 s and 72 °C for 20 s. The relative lncRNA and mRNA expression levels were calculated using the 2−ΔΔCt method. The expression levels of all targets were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer information could be found in
GO analysis and KEGG pathway analysis
GO analysis and KEGG pathway analysis were performed to investigate the roles of differentially expressed mRNAs. Briefly, GO analysis was conducted to illustrate the genetic regulatory networks in accordance with three integrated domains (molecular function, biological process, and cellular component). The number of differentially expressed genes included in each GO term was counted, and Hypergeometric cumulative distribution function was used to calculate the enrichment of functional terms. The smaller the p-value, the more enriched the differentially expressed gene was in the GO term. The top 20 terms were sorted according to the p-value. Pathway analysis of differentially expressed genes was performed using the KEGG database, and Hypergeometric cumulative distribution function was used to calculate the enrichment of pathway terms. Again, the top 20 terms were sorted according to the p-value.
Construction of the coexpression network
Coexpression networks were created to identify the interactions among lncRNAs and the protein-coding genes involved in nanotubular topography-induced biological and molecular pathways. On the basis of Guttman et al18–20, lncRNAs that participate in special biological pathways are regulated by critical transcription factors (TFs) of those pathways. In order to determine whether the lncRNAs could possess trans-regulating functions, we compared the mRNAs that are coexpressed with lncRNAs with particular TFs regulated mRNAs. To investigate the mutual regulatory relationships among miRNA, lncRNA, and mRNA, we performed miRNA–lncRNA–mRNA coexpression network analysis based on Pearson’s correlation coefficient. Coexpression relationships with more than three shared miRNAs at p<0.05 were selected to build the miRNA–lncRNA–mRNA interaction network. The networks were constructed using the hypergeometric cumulative distribution function in MATLAB 2012b (The MathWorks, Natick, USA). The graphs were generated in Cytoscape 3.01 (Agilent Technologies).
Statistical analysis
All data were shown as the mean ± standard deviation and were performed using SPSS 23.0 (SPSS Inc., Chicago, USA) statistics software. Student’s t-test was applied to comparisons between two groups. A p-value <0.05 was considered statistically significant.
Results
Material characterization
The morphology of the nitinol-based nanotubular coatings was characterized by scanning electron microscope and is shown in Figure 1A and B. The typical outer diameter of the hollow nanotubes fabricated in this study was ~50 nm. The wettability of the implant surface plays a significant role in cell adhesion,21 so the surface hydrophilicity was evaluated by the measurement of the water contact angle. As shown in Figure 1C, the contact angle of the water on the planar nitinol surface without anodic oxidation was 77.8±0.7 °. After anodic oxidation and thermal treatment, the contact angle of the water decreased to 12.7±0.2 °, indicating that the nitinol-based nanotubular surface was more hydrophilic than the planar nitinol surface.
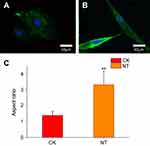
Cytoskeletal organization and phenotypic expression
In general, cells undergo an adhesion process that involves substrate attachment, spreading, and cytoskeletal development.22 We investigated the cytoskeletal organization of HCAECs grown on planar nitinol (CK) and nanotubular coatings (NT) by staining the cells with phalloidin and DAPI. We then compared the organization of the actin structures on the nanotubular coatings with those on the planar nitinol surfaces. Confocal microscopy showed that the cells on the flat surfaces were spread out, however, cells on nanotubular coatings had elongated morphologies (Figure 2A and B). The aspect ratio of cells in the NT group was significantly higher than that of cells in the CK group (Figure 2C). It has been reported that elongation can be a response to mechanical forces induced by topographical cues.23 Moreover, a higher migration speed has been observed in elongated cells compared with spread cells.8 Based on this evidence, nanotubular topography may facilitate the migration of HCAECs, resulting in rapid re-endothelialization, which is pivotal for wound healing after device implantation.
The phenotypic expression of the endothelial cell marker CD31, the focal adhesion protein vinculin, and the mature endothelial cell marker vWF was confirmed in cells on two different substrate surfaces (
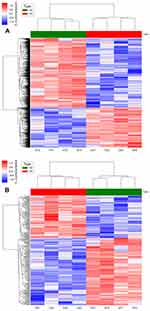
Differentially expressed lncRNAs
Microarray analysis was performed to investigate the expression levels of lncRNAs in HCAECs on nanotubular coatings compared with those of lncRNAs in cells on control planar nitinol. In total, we identified 1085 significantly differentially expressed lncRNAs in the HCAECs cultured on nanotubular coatings; 687 were upregulated, while 398 were downregulated (≥2.0-fold, p<0.05). The 40 most significantly differe-ntially expressed lncRNAs can be found in
Differentially expressed mRNAs
Microarray analysis identified 227 mRNAs that were significantly differentially expressed in the cells cultured on the nanotubular coatings but not in the cells cultured on control planar nitinol; 136 were upregulated, while 91 were downregulated (≥2.0-fold, p<0.05). The 40 most significantly differentially expressed mRNAs can be found in
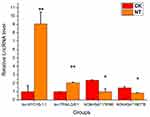
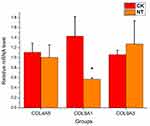
Validation of the microarray data using qRT-PCR
qRT-PCR was performed to confirm the microarray analysis findings. Four differentially expressed lncRNAs (lnc-MYO1B-1:1, lnc-TRIML2-8:1, NONHSAT178380, and NONHSAT190776) were randomly selected for validation. The expression levels of these lncRNAs are shown in Figure 5. In accordance with the microarray analysis, the results of qRT-PCR demonstrated that lnc-MYO1B-1:1 and lnc-TRIML2-8:1 were upregulated in the NT group compared with the CK group, while NONHSAT178380 and NONHSAT190776 were downregulated.
Microarray-based GO analysis and pathway analysis
Functional analyses for differentially expressed mRNAs were performed by GO and pathway analyses. In this study, we found that some of the most important GO terms were related to cell migration. For example, the most enriched GO terms in the upregulated transcripts were “exocyst localization” (ontology aspect: biological process,
Construction of the coexpression network
A gene coexpression network was constructed with the differentially expressed lncRNAs and mRNAs with absolute values of the Pearson correlation coefficient >0.9 and p<0.05 (
 | Figure 7 Top-ranking TFs associated with the differently expressed lncRNAs according to p-value. |
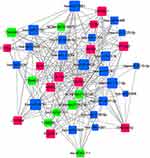 | Figure 10 miRNA-lncRNA-mRNA coexpression network. The red rectangles represent the mRNAs, the blue rectangles represent the miRNAs, and the green rectangles represent the lncRNAs. |
Discussion
In the present study, we investigated, for the first time, the altered expression pattern of lncRNAs in HCAECs in response to nitinol-based nanotubular coatings using microarray analysis. Also, potential roles and molecular networks of differentially expressed mRNAs and lncRNAs were predicted. In total, 1085 lncRNAs and 227 mRNAs were differentially expressed owing to the nitinol-based nanotubular coatings.
To gain insight into the potential functions of the differentially expressed mRNAs, GO and pathway analyses were performed. The most significantly altered categories identified by GO analysis were those involving the plasma membrane and exocyst-related processes. It has been shown that the exocyst interacts with Rho GTPases resulting in cell polarity and the activity of the cytoskeleton.25 In addition to regulation by the cytoskeleton, integrin activation is also regulated by small GTPases.26 Integrins are the major cell-ECM adhesion receptors. They are also mechanotransducers that can modulate cytoskeletal remodeling through the regulation of Rho family small GTPases.26 It is suggested that the nanotubular topography influences the interaction between integrins and Rho GTPases in HCAECs through plasma membrane and exocyst-related processes, leading to the change of cytoskeleton organization. In addition, GO analysis identified the most important upregulated molecular functions as G-protein coupled receptor activity, calmodulin-dependent protein kinase activity and calcium ion binding, all of which have previously been implicated in mechanotransduction.27 It has been reported that Ca2+ signaling can activate G-proteins via a mechanical signal.28 G-proteins can also regulate mechanotransduction and activation of specific G proteins may modulate the activity of specific Rho GTPases,26 which can interact with exocyst participating in exocyst-related processes mentioned above. Previous studies have suggested that increased roughness on stainless steel surfaces may improve the expression of cell adhesion molecules in ECs such as E-selectin and VCAM-1;29 the most highly correlated pathways that we identified were those related to ECM–receptor interaction and cell adhesion molecules. Therefore, we speculate that the nanotubular topography can affect the cell adhesion molecules located on the surface of the cell membrane, and regulate the behavior and function of endothelial cells by downstream G–protein coupled receptors and Ca2+ signaling. However, all of these speculations need to be studied in future work.
Among the top-ranking transfactors associated with the differentially expressed lncRNAs, TBP and TAF1 are the most important. It has been reported that TBP levels regulate the expression of VEGFA, which is a rate-limiting growth factor for angiogenesis. Furthermore, improvement of cellular TBP levels contribute to VEGFA expression and secretion involved in enhancing cell migration.24 Whether it is possible for nanotubular topography to regulate the behavior of endothelial cells via the expression of TBP and VEGFA is also worthy of further exploration. We also identified another significant transfactor, E2F4, that was associated with the differently expressed lncRNAs. E2F4 belongs to the E2F family, which is a series of transcription factors involved in cell proliferation in response to extra- and intra-cellular signals.30 It has been shown that E2F3 promotes the proliferation of vascular smooth muscle cells, resulting in increased intimal hyperplasia, whereas E2F4 inhibits this pathological response.31 Therefore, further investigation of the potential roles of E2F4-associated differently expressed lncRNAs in intimal hyperplasia inhibition is warranted. Overall, we speculate that the transcription factors TBP and TAF1 in the nucleus regulate the expression of certain genes contributing to changes in cell function. The functions of TBP and TAF1 involved in EC responses to substrate topography need to be proved by further experiments.
However, one limitation of this study is that the effect of shear stress on blood flow was not explored. Blood flow shear stress is also an important factor influencing endothelial cells after stent implantation.Shear stress has an intense effect on the function and behavior of vascular endothelial cells, so future research should explore the changes in lncRNA expression when the cells are subjected to shear stress and substrate topography together. In addition, it has been proved that smooth muscle cells are also an important cell type after stenting, so we will focus on such cells in further research. By comparing the gene expression between endothelial cells and smooth muscle cells, surfaces of vascular stents that are more favorable to endothelial cell proliferation and function can be designed, and the function of vascular stents can be further promoted.
Conclusion
In this work, a microarray analysis was performed to confirm the 1085 lncRNAs differentially expressed in HCAECs in response to nitinol-based nanotubular coatings. Potential roles and molecular networks of these differentially expressed mRNAs and lncRNAs were also predicted. Extracellular matrix receptor interactions and cell adhesion molecules play critical roles in the sensing of nitinol-based nanotubular coatings by HCAECs. TBP and TAF1 are important molecules in EC responses to substrate topography, but their functions involved in EC responses to nanotubular topography need to be proved by further experiments. Identifying the lncRNAs and networks involved in the responses to nanotopographical cues may be crucial in understanding how cells respond to nanotopography. Furthermore, revealing how cells and topography interact can improve the design of future medical devices.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81600148, 51621003, U1607110) and the Beijing Municipal Commission of Education Foundation (KZ201610005002).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Forouzanfar MH, Moran AE, Flaxman AD, et al. Assessing the global burden of ischemic heart disease, part 2: analytic methods and estimates of the global epidemiology of ischemic heart disease in 2010. Glob Heart. 2012;7(4):331–342. doi:10.1016/j.gheart.2012.10.003
2. Di NM, Middeldorp S, Büller HR. Direct thrombin inhibitors. New England Journal of Medicine. 2005;353(10):1028. doi:10.1056/NEJMra044440
3. Avci-Adali M, Ziemer G, Wendel HP. Induction of EPC homing on biofunctionalized vascular grafts for rapid in vivo self-endothelialization–a review of current strategies. Biotechnol Adv. 2010;28(1):119–129. doi:10.1016/j.biotechadv.2009.10.005
4. Castellanos MI, Guillem‐Marti J, Mas‐Moruno C, et al. Cell adhesive peptides functionalized on CoCr alloy stimulate endothelialization and prevent thrombogenesis and restenosis. J Biomed Mater Res A. 2017;105(4):973. doi:10.1002/jbm.a.35988
5. Bettinger CJ, Langer R, Borenstein JT. Engineering substrate topography at the micro- and nanoscale to control cell function. Angewandte Chemie. 2010;48(30):5406–5415. doi:10.1002/anie.200805179
6. Tan CH, Muhamad N, Abdullah MMAB Surface topographical modification of coronary stent: a review.
7. Peng L, Eltgroth ML, Latempa TJ, Grimes CA, Desai TA. The effect of TiO2 nanotubes on endothelial function and smooth muscle proliferation. Biomaterials. 2009;30(7):1268–1272. doi:10.1016/j.biomaterials.2008.11.012
8. Brammer KS, Oh S, Gallagher JO, Jin S. Enhanced cellular mobility guided by TiO2 nanotube surfaces. Nano Lett. 2008;8(3):786–793. doi:10.1021/nl072572o
9. Zhong S, Luo R, Wang X, et al. Effects of polydopamine functionalized titanium dioxide nanotubes on endothelial cell and smooth muscle cell. Colloids Surf B Biointerfaces. 2014;116(5):553–560. doi:10.1016/j.colsurfb.2014.01.030
10. Lee PP, Cerchiari A, Desai TA. Nitinol-based nanotubular coatings for the modulation of human vascular cell function. Nano Lett. 2014;14(9):5021–5028. doi:10.1021/nl501523v
11. Archer K, Broskova Z, Bayoumi AS, et al. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. 2015;16(10):23651–23667. doi:10.3390/ijms161023651
12. Gomes CPC, Spencer H, Ford KL, et al. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids. 2017;8(C):494–507. doi:10.1016/j.omtn.2017.07.014
13. Boon RA, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67(10):1214–1226. doi:10.1016/j.jacc.2015.12.051
14. Zhou T, Ding JW, Wang XA, Zheng X-X. Long noncoding RNAs and atherosclerosis. Atherosclerosis. 2016;248:51–61. doi:10.1016/j.atherosclerosis.2016.02.025
15. Wang N, Li H, Lü W, et al. Effects of TiO nanotubes with different diameters on gene expression and osseointegration of implants in minipigs. Biomaterials. 2011;32(29):6900–6911. doi:10.1016/j.biomaterials.2011.06.023
16. Lv L, Liu Y, Zhang P, et al. The nanoscale geometry of TiO2 nanotubes influences the osteogenic differentiation of human adipose-derived stem cells by modulating H3K4 trimethylation. Biomaterials. 2015;39(Complete):193–205. doi:10.1016/j.biomaterials.2014.11.002
17. Wenwen L, Penglei S, Su C, et al. Synthesis of TiO2 nanotubes with ZnO nanoparticles to achieve antibacterial properties and stem cell compatibility. Nanoscale. 2014;6(15):9050. doi:10.1039/C4NR01531B
18. Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding Rnas in mammals. Nature. 2009;458:223–227. doi:10.1038/nature07672
19. Mitchell G, Julie D, Carey BW, et al. lncRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295. doi:10.1038/nature10398
20. Alexander S, Johannes KS, Tobias M, et al. Next-generation RNA sequencing reveals differential expression of MYCN target genes and suggests the mTOR pathway as a promising therapy target in MYCN-amplified neuroblastoma. Int J Cancer. 2012;132(3):E106–E115. doi:10.1002/ijc.27787
21. Wilson CJ, Clegg RELeavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11(2):1–18. doi:10.1089/ten.2005.11.1
22. Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–4415.
23. Sato M, Ohashi T. Biorheological views of endothelial cell responses to mechanical stimuli. Biorheology. 2005;42(6):421.
24. Sas J, Lin JJ, Walkey CJ, Leathers MP, Coarfa C, Johnson DL. Elevated TATA-binding protein expression drives vascular endothelial growth factor expression in colon cancer. Oncotarget. 2017;8(30):48832–48845. doi:10.18632/oncotarget.16384
25. Ory S, Gasman S. Rho GTPases and exocytosis: what are the molecular links? Semin Cell Dev Biol. 2011;22(1):27–32. doi:10.1016/j.semcdb.2010.12.002
26. Song L, Huang NF, Steven H. Mechanotransduction in endothelial cell migration. J Cell Biochem. 2010;96(6):1110–1126.
27. Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10(1):63. doi:10.1038/nrm2597
28. Plank MJ, Wall DJN, David T. Atherosclerosis and calcium signalling in endothelial cells. Prog Biophys Mol Biol. 2006;91(3):287–313. doi:10.1016/j.pbiomolbio.2005.07.005
29. Mclucas E, Moran MT, Rochev Y, Carroll WM, Smith TJ. An investigation into the effect of surface roughness of stainless steel on human umbilical vein endothelial cell gene expression. Endothelium J Endoth Cell Res. 2006;13(1):35. doi:10.1080/10623320600660185
30. Hsu J, Sage J. Novel functions for the transcription factor E2F4 in development and disease. Cell Cycle. 2016;15(23):3183–3190. doi:10.1080/15384101.2016.1234551
31. Giangrande PH, Zhang J, Tanner A, et al. Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia. Proc Natl Acad Sci. 2007;104(32):12988–12993. doi:10.1073/pnas.0704754104
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.