Back to Journals » Drug Design, Development and Therapy » Volume 12
Integration of microRNA–mRNA profiles and pathway analysis of plant isoquinoline alkaloid berberine in SGC-7901 gastric cancers cells
Authors Yang YH , Zhang N, Li KD, Chen J, Qiu L, Zhang JF
Received 3 November 2017
Accepted for publication 22 January 2018
Published 28 February 2018 Volume 2018:12 Pages 393—408
DOI https://doi.org/10.2147/DDDT.S155993
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Manfred Ogris
Yanhong Yang,1 Na Zhang,2 Kundong Li,2 Juan Chen,2 Lang Qiu,1 Jufeng Zhang2
1The First Affiliated Hospital, School of Clinical Medicine, Guangdong Pharmaceutical University, Guangzhou, People’s Republic of China; 2School of Life Science and Biopharmacology, Guangdong Pharmaceutical University, Guangzhou Higher Education Mega Center, Guangzhou, People’s Republic of China
Purpose: Berberine (BBR) is a traditional Chinese medicine normally used for gastroenteritis, and recent research found that it could fight against tumors. In this study, we focused on integrating miRNA sequencing and RNA sequencing of SGC-7901 gastric cancer cells treated by BBR to elucidate their underlying mechanisms.
Materials and methods: WST-1 assay and flow cytometry were used to check the effects of BBR on SGC-7901. miRNA sequencing and RNA sequencing were used to establish the miRNA and mRNA profiles of BBR-treated SGC-7901.
Results: The results showed that BBR could inhibit the proliferation of SGC-7901 cells and induce G1 arrest in cell cycle phase and apoptosis. A total of 1,960 upregulated genes and 4,837 downregulated genes were identified in the RNA sequencing and 347 upregulated and 93 downregulated miRNAs in the miRNA sequencing. A total of 78 novel miRNAs were also found. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis showed that the genes were related to pathways in cancer and metabolism. We also analyzed the miRNA–mRNA network of genes grouped into cell cycle, apoptosis, inflammation, metabolism, cell junction, acetylization process, TGF-β pathway, and Wnt signaling pathway.
Conclusion: BBR could inhibit the proliferation of SGC-7901 cells and induce apoptosis. Integrated analysis of microRNA–mRNA profiles is a promising approach to validate gene expression patterns associated with malignant phenotype and study the mechanisms of anticancer.
Keywords: berberine, gastric cancer, RNA sequencing, microRNA sequencing, integrated analysis
Introduction
Berberine (BBR) is a bioactive alkaloid that can be isolated from several Chinese herbs, such as Huang lian (Rhizoma Coptidis).1 BBR is often used for the treatment of diarrhea and gastroenteritis, but many other pharmacological and biochemical effects of the alkaloid have been found recently. It has been reported that BBR is effective against a number of diseases including hyperlipidemia, diabetes, metabolic syndrome, and Alzheimer’s disease,2–5 and a growing number of reports show that BBR has anticancer effects.6,7
However, the diversity of BBR targets that mediate these pharmacological activities has not been well understood yet. It has been reported that BBR directly regulates gene expression by targeting TATA boxes in transcriptional regulatory regions as well as the poly(A) tail at the mRNA terminus, and the final outcome of gene regulation by BBR depends on the structure of the individual gene.8 Recent researches have shown that BBR could also regulate the expression of miRNAs in several diseases including cancers and diabetes.9,10 It was reported that treatment of 3T3-L1 cells with BBR inhibited cell viability and differentiation in a dose- and time-dependent manner, and significantly increased the expression levels of miRNA-27a and miRNA-27b.11 In human hepatocellular carcinoma cells treated by BBR, miR-23a was upregulated and the upregulation of miR-23a by BBR is p53 dependent.12 It was also reported that treatment of human U266 multiple myeloma cell line with BBR suppressed cell proliferation and IL-6 secretion in a dose- and time-dependent manner. Meanwhile, BBR dose dependently induced ROS generation, G2/M phase arrest, and apoptosis in U266 cells, and decreased the levels of miR-21 and Bcl-2.13
For the effects of BBR on gastric cancer, Zhang et al reported that BBR or d-limonene alone could inhibit the growth of MGC803 cells in a dose- and time-dependent manner, and the combination of BBR and d-limonene showed more remarkable effects on MGC803 cells by cell cycle arrest, ROS production, and apoptosis induction through the mitochondria-mediated intrinsic pathway compared with drugs used singly.14 You et al reported that BBR treatment significantly reversed cisplatin sensitivity and induced caspase-dependent apoptosis in cisplatin-resistant SGC-7901 and BGC-823 cells by modulating the miR-203/Bcl-w apoptotic axis.15
Although BBR has anticancer effects on gastric cancer cells, the mechanisms are not clear yet. More advanced tools with a large scale of data are required for the deep interpretation of the problems. Here, in this study, we performed an integrative analysis of the miRNA–mRNA expression profiles in BBR-treated SGC-7901 gastric cancer cells by high-throughput miRNA sequencing and mRNA sequencing technologies. We have integrated miRNA and mRNA expression profiles and regular laboratory analysis to understand the effects of BBR on gastric cancer and the mechanisms. Although BBR is an “old” medicine, it may become a novel agent for the treatment of gastric cancer.
Materials and methods
Cell culture
The human gastric cancer cell line SGC-7901 was purchased from American Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco; Thermo Fisher Scientific Waltham, MA, USA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) at 37°C in a humidified atmosphere containing 5% CO2. When needed, cells were treated with BBR (Beyotime Biotechnology Company, Shanghai, People’s Republic of China, molecular weight 371.82, purified by high-performance liquid chromatography) at different concentrations.
WST-1 assay
SGC-7901 cells were plated into a 96-well plate at a density of 4×103/well and treated with BBR for 12 and 24 h at different concentrations. Then, cells were treated with WST-1 and culture was continued for 2.5 h according to the protocol of the WST-1 Assay Kit (Beyotime Biotechnology Company). Finally, the observation density was read at 450 nm by a plate reader (BioTek Epoch, Winooski, VT, USA). The percentage of proliferation inhibition was calculated using the following formula: (1–the OD value of the BBR-treated group/the OD value of the control group) ×100%. The experiment was repeated at least three times.
Cell cycle and apoptosis detection by flow cytometry
SGC-7901 cells were seeded in six-well plates at a density of 1×105/well and incubated with BBR at different concentrations for 24 h. Cells were harvested and washed with phosphate-buffered saline, and fixed with 70% ethanol at 4°C overnight. After staining with propidium iodide (PI) solution for 30 min, cells were collected on a FACScan flow cytometer (Accuri™ C6 Flow cytometer; BD Biosciences, San Jose, CA, USA) equipped with a 488 nm argon laser and analyzed using ModFit LT. Cells for detection of apoptosis were treated with BBR at different concentrations for 24 h, and stained with Annexin-V and PI (BD Pharmingen™; BD Biosciences) according to the manufacturer’s instruction. Flow cytometry analysis was also performed by FACScan flow cytometer and analyzed by FlowJo 7.6.
Library construction and miRNA sequencing analysis
MiRNA isolation, library construction, and miRNA sequencing were carried out by Gene Denovo Biotechnology Company (Guangzhou, People’s Republic of China). Total RNA was extracted by TRIzol reagent, and the RNA molecules in a size range of 18–30 nt were enriched by polyacrylamide gel electrophoresis. Then the 3′ adapters were added and the 36–44 nt RNAs were enriched. The 5′ adapters were then ligated to the RNAs as well. The ligation products were reverse transcribed by PCR amplification, and the 140–160 bp size PCR products were enriched to generate a cDNA library. The cDNA library was sequenced by Illumina HiSeqTM2500, (San Diego, CA, USA) following the vendor’s recommended protocol.
Library construction and RNA sequencing analysis
RNA isolation, library construction, and RNA sequencing were carried out by Gene Denovo Biotechnology Company. After total RNA was extracted, mRNA was enriched by Oligo (dT) beads, and the enriched mRNA was fragmented into short fragments and reverse transcribed into cDNA with random primers. Then the cDNA fragments were purified with QiaQuick PCR extraction kit, poly(A) added and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeqTM2500.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis
Enrichment analysis of the genes was conducted with Gene Ontology (GO) (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www.genome.jp/kegg/). A heatmap chart was drawn by transforming the normalized data clustering using the gplots heatmap.2 of R program.
Statistical analysis
Statistical differences were determined by using SPSS 23.0 software. A P-value <0.05 was considered to be significant.
Results
Effect of BBR on the proliferation of SGC-7901 cells detected by WST-1 assay
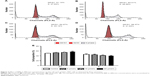
BBR at concentrations of 2.5, 5, 10, 20, and 30 μM was used to treat SGC-7901 cells for 12 and 24 h, respectively, and the results showed that BBR could inhibit the proliferation of SGC-7901 cells (Figure 1). BBR could still inhibit the proliferation of SGC-7901 cells at 48 h (Figure S1).
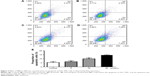
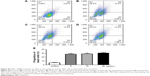
Effect of BBR on the cell cycle and apoptosis of SGC-7901 cells detected by flow cytometry
SGC-7901 cells were treated with BBR at different concentrations for 24 h, and the effect of BBR on the cell cycle and apoptosis of SGC-7901 cells was detected by flow cytometry. It was shown that the cells were accumulated in G1 phase, and less cells entered S phase (Figure 2). BBR could also induce the apoptosis of SGC-7901 cells, and the apoptosis was more obvious with the higher concentration (Figure 3). When the time of SGC-7901 cells treated by BBR was prolonged to 48 h, the number of dead cells was significantly increased in BBR-treated SGC-7901 cells compared to the control group (Figure S2), but there was no difference in apoptosis between the BBR groups and the control group.
Analysis of gene expression profile of RNA sequencing
According to the sequence data, 28,214,592 and 26,977,858 clean reads were identified in the control group and the BBR-treated group, respectively. In the control group, high-quality reads were 27,371,360 (97.01% in total) and in the BBR-treated group, high-quality reads were 26,319,510 (97.56% in total). Here, 1,960 upregulated genes and 4,837 downregulated genes were identified in the BBR-treated SGC-7901 cells compared to the control group (Figure 4).
To understand the functions of differentially expressed genes, all the differentially expressed genes were mapped to terms in the GO database and compared with the background (Figure 5). The GO has 3 ontologies, which are “biological process”, “cellular component”, and “molecular function”. In the “biological process” part, most genes were related to “cellular process”, “metabolic process”, and “single-organism process”, but few genes were related to “cell killing” and “reproductive process”. In the “cellular component” part, most genes were related to the “cell part” and “organelle part”, and few genes were related with “cell junction” and “extracellular matrix”. In the “molecular function” part, most genes were related with “binding” and “catalytic activity”.
  | Figure 5 The presentation of GO classification of the mRNA expression profile. |
The KEGG assignments were used to classify functional annotations of the identified differentially expressed genes to further understand the biological functions (Figure 6). Top 20 pathways were shown in the result, including regulation of autophagy, mTOR signaling pathway, MAPK (mitogen-activated protein kinase) signaling pathway, insulin resistance, microRNAs in cancer, AMPK (AMP-activated protein kinase) pathway, pathways in cancer, and pathways related to metabolism.
  | Figure 6 The presentation of KEGG assignments of differentially expressed genes. |
Analysis of miRNA profile of miRNA sequencing
The result of the miRNA sequencing showed that 11,551,248 and 10,360,439 clean reads were identified in the control group and the BBR-treated group, respectively. In the control group, high-quality reads were 11,135,724 (96.40% in total), and in the BBR-treated group, high-quality reads were 9,987,143 (96.40% in total). Here, 347 upregulated and 93 downregulated miRNAs were identified in the BBR-treated SGC-7901 cells compared to the control group (Figure 7).
  | Figure 7 The miRNA expression profile of SGC-7901 cells treated by BBR. |
The target genes of the upregulated or downregulated miRNAs were mapped to terms in the GO database (Figure 8). In the “biological process” part, most genes were related to “cellular process”, “metabolic process”, and “single-organism process”. In the “cellular component” part, most genes were related to the “cell”, “cell part”, and “organelle part”. In the “molecular function” part, most genes were related with “binding” and “catalytic activity”.
  | Figure 8 The presentation of GO classification of the upregulated or downregulated miRNA target genes. |
The top 20 pathways of the KEGG assignments of the target genes of differentially expressed miRNAs are shown in Figure 9, including Ras signaling pathway, Jak-STAT signaling pathway, P13K-Akt signaling pathway, Rap1 signaling pathway, insulin signaling pathway, AMPK signaling pathway, cAMP signaling pathway, and Oxytocin signaling pathways.
  | Figure 9 The presentation of KEGG assignments of the upregulated or downregulated miRNA target genes. |
Novel miRNAs and target genes
According to the results of miRNA sequencing, 78 novel miRNAs were found, 71 found in the control group and 55 found in the BBR-treated group. KEGG showed that the pathways of the target genes of differently expressed miRNAs mainly focused to “calcium pathway”, “PI3K-Akt signaling pathway”, “Ras signaling pathway”, “cAMP signaling pathway”, and “pathways in cancer” (Figure 10).
  | Figure 10 The presentation of KEGG assignments of the upregulated or downregulated novel miRNA target genes. |
Analysis of miRNA–mRNA network
Based on the result that BBR could inhibit the SGC-7901 cells, the gene expression profile was further analyzed in detail. Genes related to cell cycle, apoptosis, inflammation, and metabolism were picked up, and their expression level and miRNA–RNA network were further analyzed. TGF-β pathway and Wnt signaling pathway were also analyzed. In addition, genes related to cell junction and acetylization were also differentially expressed in the BBR-treated group. The merged heatmap is shown in Figure 11, and the merged miRNA–RNA relation network is shown in Figure 12. Several novel anticancer pathways were also found in BBR-treated SGC-7901 cells, such as Hippo pathway, Notch pathway, and FoxO pathway. The merged heatmap and miRNA–RNA relation network of the three pathways are shown in Figures S3 and S4, respectively.
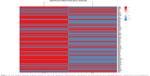  | Figure 11 The merged heatmap of genes related to cell cycle, apoptosis, inflammation, metabolism, TGF-β pathway, Wnt signaling pathway, cell junction, and acetylization. |
  | Figure 12 The merged miRNA–RNA relation network of genes related to cell cycle, apoptosis, inflammation, metabolism, TGF-β pathway, Wnt signaling pathway, cell junction, and acetylization. |
Discussion
Gastric cancer is the fourth most common cancer and the second leading cause of cancer deaths worldwide. Although its incidence and mortality have been declining due to the decline in the prevalence of Helicobacter pylori infection and the improvements in food preservation and diet, the burden of gastric cancer remains very high in several countries in Asia, including China.16 Most of the chemotherapy drugs for gastric cancer are cytotoxic, which have serious adverse reactions and directly affect the curative effect of gastric cancer. It is one of the effective ways to treat gastric cancer by looking for drugs from natural plants with obvious anti-tumor activity but little side effects.
Our data displayed in this paper demonstrated that BBR could inhibit the proliferation of SGC-7901 gastric cancer cells and induce G1 phase arrest and apoptosis. The KEGG analysis of the RNA sequencing showed that BBR was involved in autophagy and mTOR and MAPK signaling pathways. Pathway analysis of the miRNA target genes included cell cycle, Ras signaling pathway, Jak-STAT signaling pathway, and P13K-Akt signaling pathway. In the gene expression profiling, we also found genes related to cell cycle and apoptosis, and genes in TGF-β pathway and Wnt signaling pathway. These data of miRNA sequencing and RNA sequencing supported our results that BBR could inhibit the proliferation and induce apoptosis of SGC-7901 cells, and provided us the clues that BBR may exert its anti-tumor activities through these pathways. Li et al reported that inactivation of p38 MAPK, extracellular signal-regulated kinase 1/2, and c-Jun N-terminal kinase by BBR contributed to the decreased proliferation and tumorigenesis of MGC-803 gastric cancer cells.17 Yi et al demonstrated that Akt-related mitochondrial pathway may contribute to the BBR-induced apoptosis in human gastric cancer cells and might represent an important molecular basis for BBR to act as an anticancer agent.18 Another research demonstrated that BBR could modulate PI3K-AKT and MAPK signaling pathways in thyroid carcinoma cells, which leads to mitochondrial apoptosis, G0/G1 cell cycle arrest and suppressive migration.19 Okubo et al showed that BBR promptly localized to the nucleus after treatment in HL-60 human promyelocytic leukemia cells and induced apoptotic cell death by activation of caspase-3 and caspase-8.20 It has been reported that BBR inhibits colon tumor formation through inhibition of Wnt/β-catenin signaling and might be a promising drug for the prevention of colon cancer.21 All the conclusions from these papers have shown that it is an effective method to study the mechanisms of BBR through high-throughput miRNA sequencing and RNA sequencing technologies.
The results of KEGG analysis also demonstrated that BBR is also involved in insulin resistance and metabolism pathways, inflammation pathways, cell junction, and acetylization. Wang et al reported that BBR was able to significantly reduce the alcohol-induced gastrointestinal injury, inhibit increases of alcohol-induced TNFα and IL-1β expression in gastrointestinal mucosa as well as their upstream signals TLR2 and TLR4, and regulate cytokines that modulate tight junctions.22 Zhao et al demonstrated that BBR and nandinine attenuated insulin resistance in adipocytes by inhibiting inflammation in an AMP-activated protein in a kinase-dependent manner, and BBR and nandinine may be used as dietary supplements for obesity treatment.23 It has also been reported that ischemia/reperfusion-induced intestinal tight junction dysfunction could be improved by BBR, thereby demonstrating the therapeutic potential of BBR for intestinal ischemia/reperfusion injury.24
Among the epigenetic changes, histone acetylation has been recognized as a fundamental process that strongly affects gene expression regulation, and disruption of this phenomenon has been linked to carcinogenesis.25 Seldom research is about the effects of BBR on acetylation and cancer. Zhang et al reported that BBR treatment could restore the expression of the L-type pyruvate kinase (L-PK) by the demethylation of L-PK promoter and the increase in acetylation levels of histone H3 and H4 around L-PK, which indicated that BBR may be a potential drug for non-alcoholic fatty liver disease and diabetes.26 It still needs further study about the effects of BBR on gastric cancer.
In this report, we have integrated regular laboratory analysis and high-throughput miRNA sequencing and RNA sequencing to study the effects of BBR on gastric cancer cells and its mechanisms. Our data showed that BBR could inhibit the proliferation of SGC-7901 cells and induce G1 arrest in cell cycle phase and apoptosis.
We have got the miRNA and mRNA expression profiles of SGC-7901 gastric cancer cells treated with BBR as well as the mechanisms involved in cell cycle, apoptosis, inflammation, metabolism, cell junction, acetylization process, TGF-β pathway, and Wnt signaling pathway. Based on the results of the mRNA expression profile, genes related to cell cycle, apoptosis, inflammation, metabolism, TGF-β pathway, Wnt signaling pathway, cell junction, and acetylization were further analyzed in the miRNA–mRNA network. We also found some novel miRNAs.
Conclusion
BBR has several pharmacological and biochemical effects, and the underlying mechanisms are still far from being completely understood. Here in this study, we reported the findings of comprehensive miRNA–mRNA investigation of BBR on gastric cancer by high-throughput sequencing technology with network pathway analysis. Our findings have illustrated the power of high-throughput sequencing for large-scale biological data sets, which may be a promising method to provide insight into the action mechanisms of BBR and help to guide its predictions.
Data availability
The RNA sequencing, miRNA sequencing, and the novel miRNAs data have been uploaded to figshare (https://figshare.com/), doi:10.6084/m9.figshare.5738577. Anyone who is interested in the study of miRNAs, BBR, cancers, and related fields can download the data for free.
Acknowledgment
This work was supported by the Guangdong Science and Technology Program (number 2014A020212303) and the Research Project of the First Affiliated Hospital of Guangdong Pharmaceutical University (number GYFY201601).
Disclosure
The authors report no conflicts of interest in this work.
References
Chen XW, Di YM, Zhang J, Zhou ZW, Li CG, Zhou SF. Interaction of herbal compounds with biological targets: a case study with berberine. Sci World J. 2012;2012:708292. | ||
Kong W, Wei J, Abidi P, et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10(12):1344–1351. | ||
Tang LQ, Wei W, Chen LM, Liu S. Effects of berberine on diabetes induced by alloxan and a high-fat/high-cholesterol diet in rats. J Ethnopharmacol. 2006;108(1):109–115. | ||
Tillhon M, Guamán Ortiz LM, Lombardi P, Scovassi AI. Berberine: new perspectives for old remedies. Biochem Pharmacol. 2012;84(10):1260–1267. | ||
Yao J, Kong W, Jiang J. Learning from berberine: treating chronic diseases through multiple targets. Sci China Life Sci. 2015;58(9):854–859. | ||
Refaat A, Abdelhamed S, Yagita H, et al. Berberine enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast cancer. Oncol Lett. 2013;6(3):840–844. | ||
Zou K, Li Z, Zhang Y, et al. Advances in the study of berberine and its derivatives: a focus on anti-inflammatory and anti-tumor effects in the digestive system. Acta Pharmacol Sin. 2017;38(2):157–167. | ||
Yuan ZY, Lu X, Lei F, et al. TATA boxes in gene transcription and poly (A) tails in mRNA stability: new perspective on the effects of berberine. Sci Rep. 2015;5:18326. | ||
Ayati SH, Fazeli B, Momtazi-Borojeni AA, Cicero AFG, Pirro M, Sahebkar A. Regulatory effects of berberine on microRNome in cancer and other conditions. Crit Rev Oncol Hematol. 2017;116:147–158. | ||
Chang W. Non-coding RNAs and berberine: a new mechanism of its anti-diabetic activities. Eur J Pharmacol. 2017;795:8–12. | ||
Wu YY, Huang XM, Liu J, et al. Functional study of the upregulation of miRNA-27a and miRNA-27b in 3T3-L1 cells in response to berberine. Mol Med Rep. 2016;14(3):2725–2731. | ||
Wang N, Zhu M, Wang X, Tan HY, Tsao SW, Feng Y. Berberine-induced tumor suppressor p53 up-regulation gets involved in the regulatory network of miR-23a in hepatocellular carcinoma. Biochim Biophys Acta. 2014;1839(9):849–857. | ||
Hu HY, Li KP, Wang XJ, et al. Set9, NF-κB, and microRNA-21 mediate berberine-induced apoptosis of human multiple myeloma cells. Acta Pharmacol Sin. 2013;34(1):157–166. | ||
Zhang XZ, Wang L, Liu DW, Tang GY, Zhang HY. Synergistic inhibitory effect of berberine and d-limonene on human gastric carcinoma cell line MGC803. J Med Food. 2014;17(9):955–962. | ||
You HY, Xie XM, Zhang WJ, Zhu HL, Jiang FZ. Berberine modulates cisplatin sensitivity of human gastric cancer cells by upregulation of miR-203. In Vitro Cell Dev Biol Anim. 2016;52(8):857–863. | ||
Ferro A, Peleteiro B, Malvezzi M, et al. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50(7):1330–1344. | ||
Li HL, Wu H, Zhang BB, Shi HL, Wu XJ. MAPK pathways are involved in the inhibitory effect of berberine hydrochloride on gastric cancer MGC 803 cell proliferation and IL-8 secretion in vitro and in vivo. Mol Med Rep. 2016;14(2):1430–1438. | ||
Yi T, Zhuang L, Song G, Zhang B, Li G, Hu T. Akt signaling is associated with the berberine-induced apoptosis of human gastric cancer cells. Nutr Cancer. 2015;67(3):523–531. | ||
Li L, Wang X, Sharvan R, Gao J, Qu S. Berberine could inhibit thyroid carcinoma cells by inducing mitochondrial apoptosis, G0/G1 cell cycle arrest and suppressing migration via PI3K-AKT and MAPK signaling pathways. Biomed Pharmacother. 2017;95:1225–1231. | ||
Okubo S, Uto T, Goto A. Berberine induces apoptotic cell death via activation of Caspase-3 and -8 in HL-60 human leukemia cells: nuclear localization and structure-activity relationships. Am J Chin Med. 2017;45(7):1497–1511. | ||
Zhang J, Cao H, Zhang B, et al. Berberine potently attenuates intestinal polyps growth in ApcMin mice and familial adenomatous polyposis patients through inhibition of Wnt signaling. J Cell Mol Med. 2013;17(11):1484–1493. | ||
Wang XP, Lei F, Du F, et al. Protection of gastrointestinal mucosa from acute heavy alcohol consumption: the effect of berberine and its correlation with TLR2, 4/IL1β-TNFα signaling. PLoS One. 2015;10:e0134044. | ||
Zhao W, Ge H, Liu K, Chen X, Zhang J, Liu B. Nandinine, a derivative of berberine, inhibits inflammation and reduces insulin resistance in adipocytes via regulation of AMP-kinase activity. Planta Med. 2017;83(3–4):203–209. | ||
Gu L, Li N, Yu W, et al. Berberine reduces rat intestinal tight junction injury induced by ischemia-reperfusion associated with the suppression of inducible nitric oxide synthesis. Am J Chin Med. 2013;41(6):1297–1312. | ||
Chrun ES, Modolo F, Daniel FI. Histone modifications: a review about the presence of this epigenetic phenomenon in carcinogenesis. Pathol Res Pract. 2017;213(11):1329–1339. | ||
Zhang Y, Chang X, Song X, et al. Berberine reverses abnormal expression of L-type pyruvate kinase by DNA demethylation and histone acetylation in the livers of the non-alcoholic fatty disease rat. Int J Clin Exp Med. 2015;8(5):7535–7543. |
Supplementary materials
  | Figure S3 The merged heatmap of genes related to Hippo pathway, Notch pathway, and FOXO pathway. |
  | Figure S4 The merged miRNA–RNA relation network of genes related to Hippo pathway, Notch pathway, and FOXO pathway. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.