Back to Journals » Drug Design, Development and Therapy » Volume 9
Insight into the mechanism of polyphenols on the activity of HMGR by molecular docking
Authors Islam B, Sharma C, Adem A, Aburawi E , Ojha S
Received 16 April 2015
Accepted for publication 27 May 2015
Published 28 August 2015 Volume 2015:9 Pages 4943—4951
DOI https://doi.org/10.2147/DDDT.S86705
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Shu-Feng Zhou
Barira Islam,1,* Charu Sharma,2,* Abdu Adem,3 Elhadi Aburawi,1 Shreesh Ojha3
1Department of Paediatrics, 2Department of Internal Medicine, 3Department of Pharmacology and Therapeutics, College of Medicine and Health Sciences, United Arab Emirates University, Al-Ain, Abu Dhabi, United Arab Emirates
*These authors contributed equally to this work
Abstract: Statins are hypolipidemic drugs that are effective in the treatment of hypercholesterolemia by attenuating cholesterol synthesis in the liver via competitive inhibition of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase. Recently, dietary changes associated with drug therapy have garnered attention as novel drugs to mitigate or ameliorate hypercholesterolemia. The present study was undertaken to observe different dietary polyphenols that can bind to the active site of HMGR and inhibit it. Results from the 12 dietary polyphenols tested reveal that polyphenols can bind to HMGR and block the binding of nicotinamide adenine dinucleotide phosphate (NADP+). We observed that the rigidity of phenolic rings prevents the polyphenols from docking to the enzyme activity site. The presence of an ester linkage between the phenolic rings in (–)-epigallocatechin-3-gallate (EGCG) and the alkyl chain in curcumin allows them to orient in the active site of the HMGR and bind to the catalytic residues. EGCG and curcumin showed binding to the active site residues with a low GRID score, which may be a potential inhibitor of HMGR. Kaempferol showed binding to HMG-CoA, but with low binding affinity. These observations provide a rationale for the consistent hypolipidemic effect of EGCG and curcumin, which has been previously reported in several epidemiological and animal studies. Therefore, this study substantiates the mechanism of polyphenols on the activity of HMGR by molecular docking and provides the impetus for drug design involving further structure–function relationship studies.
Keywords: polyphenols, HMG-CoA, EGCG, curcumin, docking, in silico
Introduction
Mortality from cardiovascular disease (CVD) is the foremost cause of death in the industrialized world and the second leading cause of death worldwide. Statistics from the World Health Organization indicate that more than 60% of morbidity, disability, and mortality in Arab countries is caused by chronic noncommunicable diseases, especially CVD, diabetes, and cancer.1
Among the several risk factors of CVD, the most important are hypertension and hypercholesterolemia. In patients with altered cholesterol homeostasis, cholesterol builds up in the coronary arteries, subsequently leading to coronary heart disease (CHD), and eventually to heart failure.2 Cholesterol is one of the critical molecules essential for the structure and function of eukaryotic lipid bilayers. Epidemiological evidence has shown a positive relationship between total cholesterol concentration and the mortality rate of CHD.3,4 In humans, the mevalonate pathway is responsible for the endogenous synthesis of cholesterol. In cholesterol synthesis, the enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) (enzyme nomenclature [EC] 1.1.1.34), which catalyses the reaction converting HMG-CoA to mevalonate, is the rate-limiting enzyme of the mevalonate pathway that produces cholesterol and other isoprenoids.5 The enzyme, HMGR, represents an important molecular target for several antihypercholesteremic drugs known as statins. Human HMGR enzyme contains 888 amino acids, with the first 339 residues as the membrane anchor domain located in the endoplasmic reticulum. A linker region is located between residues ranging from 340 to 449, while the catalytic domain, from residues ranging from 450 to 888, resides in the cytoplasm.6 The structure of the catalytic portion of human HMGR consists of a proteinic tetramer containing four actives sites formed by residues of two monomers. This active site is characterized by the so-called cis-loop (residues 682–694), which is involved in the reduction of the substrate HMG-CoA.
Statins remain the drugs of choice for the treatment of dyslipidemia in patients with CHD or substantial risk factors for CHD.7,8 However, long-term use of statins has been reported to lead to the development of type 2 diabetes mellitus and autoimmune diseases.9,10 Therefore, alternate agents with lower toxicity and wide tolerance are needed and continue to be explored for future therapeutics.
Interest in dietary fats and their role in the development of CVD have spurred a considerable amount of research in the past three decades. The reduced risk of CVD in populations with more polyphenol intake, despite a high-cholesterol and high-fat diet, constitute the French paradox, which is indicative of their beneficial effects in the maintenance of cardiovascular health owing to the multiple benefits of polyphenolic compounds.11–13 In relation to drug discovery and development, the identification of drug targets is an important step, and HMGR represents one of the major therapeutic targets to regulate cholesterol biosynthesis. Synthetic, statin-like compounds that include an HMG-like moiety have shown significant hypocholesterolemic activity. However, there are other agents without a structural HMG-like moiety that are known to inhibit HMGR, such as cholestin, diosgenin, β-sitosterols, and lycopene, among many others.
Several of the polyphenolic compounds, such as tea catechins (mainly gallate ester derivatives), have been tested successfully both in vitro and in vivo as cholesterol-lowering agents, apart from their antioxidant activities. The evidence indicates that polyphenols exert concerted, multiple-level action involving the upregulation of the low-density lipoprotein receptor, the reduction of cholesterol absorption, and the modulation of both synthetic and metabolic pathways.14 Considering the complexity of action and the short half-life of HMGR, the direct effect on HMGR activity is very difficult to recognize and isolate. The literature supporting a physiologically significant inhibition of the key rate-limiting enzyme, HMGR, in lipid metabolism by polyphenols is limited for mechanistic insights into their therapeutic use.15–17
Therefore, the present study was designed to determine whether the polyphenols can reduce cholesterol biosynthesis by inhibiting HMGR, an important therapeutic target. Revealing polyphenols as novel agents similar to the statins reasonably represents HMGR as a viable therapeutic target in new drug discovery and development. Several previous studies have demonstrated that the consumption of dietary polyphenols lowers cholesterol levels and improves cardiovascular health in humans. However, the precise pharmacological and molecular mechanism for their activity on HMG-Co reductase has not been investigated extensively. We have chosen to investigate 12 polyphenolic compounds that are the common component of diets and traditional medicines across the world. The docking studies were performed to screen the binding site and binding energy of polyphenols and to provide a mechanistic insight into the pharmacological effects and therapeutic benefits. We have chosen fluvastatin as a standard statin, which is the first synthetic HMG-Co inhibitor to be developed and used in the management of dyslipidemia in the primary and secondary prevention of CVD.
Materials and methods
Receptor protein structure
The three-dimensional (3D) coordinates of the crystal structure of HMGR in complex with fluvastatin (Protein Data Bank [PDB] ID: 1HWI) was used for docking (San Diego Supercomputer Center, University of California, San Diego, CA, USA). We used Chimera (UCSF Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco, CA, USA) and the docking program DOCK6 (UCSF DOCK Resource, University of California, San Francisco, CA, USA) to perform this study.18,19
Ligand structure
The 3D structure for eriodictyol (PDB ID: 2NNL), kaempferol (PDB ID: 3QWH), catechin (PDB ID: 4C94), quercetin (PDB ID: 2UXH), naringenin (PDB ID: 2UXU), apigenin (PDB ID: 4HKK), (–)-epigallocatechin-3-gallate (EGCG) (PDB ID: 2KDH), luteolin (PDB ID: 4HKN), and myricetin (PDB ID: 4GDR) were extracted from the ligand-bound protein structure of the PDB database. The two-dimensional (2D) structure of baicalein, curcumin, and tetrahydrocurcumin (THC) were made using ChemDraw software (Cambridgesoft, MA, USA). The 2D structures were converted to 3D using the PRODRG 2 SERVER (http://davapc1.bioch.dundee.ac.uk/prodrg/).20 These 3D ligand molecules were used individually on DOCK6 for docking on the HMGR enzyme. The coordinates of nicotinamide adenine dinucleotide phosphate (NADP+) were taken from PDB ID: 1DQA.
Molecular docking
The crystal structure of HMGR was taken from the PDB (PDB ID: 1HWI). The protein coordinates were used after removing the solvent and ligand molecules. The hydrogen atoms were added to the protein and it was then minimized using the AMBER 99SB (AMBER Molecular Dynamics, University of California, San Francisco, CA, USA) force field. A molecular surface of the protein was generated with dimethyl sulfate. The spheres from the receptor HMGR were generated using the SPHGEN program of DOCK6. A grid for grid-based scoring was generated using the GRID program of DOCK. The default parameters and grid spacing of 0.25 were used in present study. The RESP charge-fitting of electrostatic potentials was completed using ANTECHAMBER module of the AMBER11 (AMBER MD Softwares, University of California, San Francisco, CA, USA) program. Flexible ligand docking was carried out using the DOCK6 program. The default values for the parameters of the anchor-and-grow algorithm of DOCK6 were used except for maximum orientations (500) and simplex anchor maximum iterations (500). The docking poses were visualized using the program Chimera. The coordinates of NADP+ on HMGR was predicted by the structure match of the HMGR enzyme bound to fluvastatin (PDB ID: 1HW1) and that bound to NADP+ (PDB ID: 1DQA) in Chimera.
DOCK scoring and ranking
Generally, docking programs attempt to simulate protein–ligand complex structures with a threshold accuracy and speed that enables pragmatic computation and analysis. The ability to predict the probable binding mode of a ligand to differentiate correct poses and eliminate incorrect ones was based on reliable scoring functions. The DOCK6 score function calculates ligand desolvation in addition to steric and electrostatic interactions between the ligand and receptor. The electrostatic interactions between the ligand and the protein are calculated from an electrostatic potential map. The best conformations of each ligand that were clustered using complete linkage analysis were saved in mol2 formats. These files were further used to apply the remaining scoring functions. During ranking, a lower score is always indicative of a higher affinity.
Results and discussion
Comparison of NADPH and flavonoids binding on HMGR
The reductive cleavage of HMG-CoA to mevalonate utilizes two molecules of nicotinamide adenine dinucleotide phosphate-oxidase (NADP[H]).8 Since flavonoids have shown promising antioxidant activity, it is of interest if they can bind to the NADP(H) binding site of HMGR. In the present study, we tested the 3D structures of 12 common dietary polyphenols to predict the binding site on HMGR. The structural formulae of these polyphenols are presented in Figure 1. It was observed that all polyphenols tested in this study can bind to the same region as the adenine ring of NADP+, and they may therefore act as inhibitors of NADP+ binding. This corroborates the fact that polyphenols are antioxidants in nature.13,16
  | Figure 1 Chemical structures of the polyphenols used in this study. |
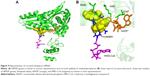
All of the polyphenols used in this docking study have been reported to possess antioxidant activity in experimental studies (Table 1). Quercetin docking to HMGR is shown as representative of polyphenol binding in Figure 2. The binding of NADP+ leads to closure of the COOH-terminal helix, moving the H866 into the hydrogen bonding distance of thiol and completing the active site.21 The blocking of NADP+ binding due to the quercetin interaction will lead to noncompletion of the active site and can inhibit the action of the HMGR enzyme. Compelling evidence from previous in vivo and in vitro studies indicates the health benefits of quercetin, such as dietary flavonoids and polyphenolic compounds in particular, in the maintenance of cardiovascular health.13,16,17,22 We predict that polyphenol binds on HMGR on the NADP+ binding site and can block the electron transfer on the substrate HMG-CoA.
Comparison of statin and polyphenol binding on HMGR
The X-ray crystal structure of human HMGR with HMG-CoA has revealed that “cis-loop” comprised of amino acid residues 682–694 forms the catalytic site of the enzyme. The residues K691, E559, and D767 of HMGR form a hydrogen bond-linked network that interacts with the carbonyl group of HMG-CoA. K691 stabilizes the negatively charged oxygen of mevaldyl-CoA. E559 and D767 are located in close proximity, which raises the pKa of E559 so that it donates a proton to mevaldehyde.21 All statins, including fluvastatin, bind to R590, K691, and D690 residues of HMGR.8 The shallow and mobile hydrophobic groove of the terminal –COOH amino acids of HMGR can accommodate the other rings of polyphenolic compounds like flavonoids. In accordance with this hypothesis, the docking studies revealed that polyphenols bind on the enzyme-active site of HMGR. The HMG-like moiety of statins binds to the catalytic residues in a similar way as HMG of HMG-CoA. As the rings of flavonoids are connected by rigid bonds, we observed that it cannot fully occupy the HMG-CoA binding site and therefore may not behave as a competitive inhibitor (Figure 2).
The supplementation of naringenin in rats has been reported to lower the plasma and hepatic cholesterol concentrations by suppressing HMGR and acetyl-coenzyme A acetyltransferase (ACAT) in rats fed a high-cholesterol diet.22 A review published on the analysis of 170 randomized clinical trials, which included 6,557 participants, concluded that no consensus can be drawn on the effect of flavonoids on the total cholesterol level of participants.23 However, we observed that EGCG, curcumin, THC, and kaempferol can bind with the active site residue (Figure 3). This is in agreement with the report that curcumin and green tea, rich in polyphenols like EGCG, have been shown to decrease cholesterol concentration.24 In agreement with this report, we observed that in polyphenols in which the phenolic rings are connected by functional groups such as –O– in EGCG and –CH2 in curcumin, binding to the enzyme-active site is stronger.
Comparing the binding of flavonoids
Differences between the flavonoid binding on the HMGR were observed based on the orientation of polyphenol rings and the side chains present on it. Among all the tested polyphenol compounds in this study, EGCG showed the best docking score and occupancy of the substrate binding site. The -gallate part of the molecule can enter the HMG-CoA binding cavity as the –OH group on the molecule forms hydrogen bonds with K691. The epigallocatechin part of EGCG interacts with the residues in the cis-loop of HMGR and –O– of ester linkage to gallate forms hydrogen bonds with R590 (Figure 3A). Recently, it has been shown that EGCG can bind to enzymes of the mevalonate pathway and inhibit cholesterol biosynthesis in vitro. The nature of binding is electrostatic, similar to the HMGR and EGCG binding shown here.25 Numerous studies have shown that catechin-rich tea extracts lower the level of serum cholesterol.24–26 EGCG has been shown to alter the expression of HMGR genes following a detailed kinetic and equilibrium study on the modulation of HMGR.16,26
Using a concerted approach involving spectrophotometric, optical biosensor and chromatographic analyses, molecular docking, and site-directed mutagenesis on the cofactor site of HMGR, the authors demonstrated that EGCG potently inhibits the in vitro activity of HMGR (Ki in the nanomolar range) by competitively binding to the cofactor site of the reductase. An earlier report based on docking experiments suggests that EGCG binds more strongly on the NADPH binding site on HMGR.16 In agreement with this study, the docking of EGCG on the enzyme-active site shows that it can cause steric hindrance to both NADPH and HMG-CoA binding. An in vitro analysis of EGCG binding on the HMG-CoA receptor shows that it inhibits the activity of the HMG-CoA receptor. The study shows that EGCG can bind on HMGR with a much higher binding affinity than other NADPH enzymes.16 Our study corroborates the previous findings and substantially aids in predicting that inhibition of HMGR activity can also possibly be due to steric hindrance caused by HMG-CoA binding.
Kaempferol and myricetin stimulate biosynthesis of cholesterol
Many experimental results suggest that curcumin is hypolipidemic and a few studies have shown a specific effect of curcumin on HMG-CoA receptor enzyme activity.17 Curcumin counteracts hyperlipidemia in animals, which otherwise would show high concentrations of circulating cholesterol and triglycerides and develop lipid-laden fatty streaks and lipid-mediated stress, including accumulation of lipids, inflammation, and increased oxidative stress within several weeks upon being fed a high-fat and/or cholesterol-rich Western diet.27 Several studies have shown that some of the effects of curcumin (eg, on cholesterol and gene expression) are similar to the effects of statins, such as lovastatin, and that curcumin mainly acts by reducing liver cholesterol biosynthesis by inhibiting, directly or indirectly, HMGR.28,30,39 In corroboration of these findings, the present docking clearly shows that curcumin can bind to the HMG-CoA site on the enzyme similar to the statins (Figures 3B and 4). Curcumin is at a distance of 3.35 Å from E559 of HMGR, which is proposed to be the proton donor for mevaldehyde.21 Previous experiments have shown that THC, which is an active metabolite of curcumin, can inhibit cholesterol synthesis comparably better than curcumin in animal models.29 It has also been also reported that the administration of THC for several weeks can significantly reduce the activity of enzyme, HMGR.30
To assess if the effect observed was due to THC binding on the HMGR active site, we performed docking of THC on the HMGR enzyme. We observed that indeed, THC can fit in the HMG-CoA binding cavity on HMGR and can profoundly inhibit the enzyme activity (Figure 3C). The GRID score of THC was markedly better than curcumin, implying that it can bind with more affinity onto the enzyme-active site (Table 2). This can be attributed to the fact that THC is more flexible when compared to curcumin, as the chain connecting the phenolic ring has single bonds, while curcumin has two double bonds in the structure. It has been clearly shown in previous studies that this conformational difference imparts rigidity to curcumin, whereas THC is twisted and flexible.31 The flexibility in THC allows better access to the enzyme-active site than curcumin, and therefore lowers the GRID score in the current DOCK-based analysis. THC has been shown to be a more potent antioxidant than curcumin in other experimental studies.32 The bioavailability of THC is also predicted to be more than curcumin.
The algorithm of the docking tools used here first uses a geometric matching algorithm to superimpose the ligand onto a negative image of the binding pocket. After, it uses an anchor-and-grow algorithm to generate conformations of the ligand on the fly within the receptor, thus allowing fine sampling of conformational space.18 In this way, the docking score reflects both the ligand fit and energy contribution due to specific interactions of the protein and ligand. It is evident from the docking results that polyphenolic compounds with a rigid three-ring structure cannot bind to the substrate binding site. However, kaempferol shows binding on the same site as statins and HMG-CoA. This is because the number and position of the –OH group of kaempferol make it most suitable to form hydrogen bond interactions and anchor on the substrate binding site (Figure 3). However, the docking score is very low compared to those for EGCG, curcumin, and THC. The docking pose indicates that kaempferol can act as an inhibitor by sterically hindering HMG-CoA binding on the enzyme. This corroborates to the findings of the epidemiological study, which predicts that among dietary flavonols and flavones, only food rich in kaempferol, like broccoli, could show slight inhibition of the cholesterol level.33
Conversely, kaempferol and myricetin have been reported to stimulate cholesterol biosynthesis, particularly within a low concentration range (from 0.1 to 10 μM), and exhibit a variable degree of inhibition at higher concentrations. These conflicting results were not further substantiated by any study, and the authors concluded that the opposing effects on cholesterol biosynthesis are exerted in an indirect manner and seem possible through differential modulation of the complex regulation of HMGR.34 A recent study suggested that treatment with kaempferol effectively prevents atherosclerosis induced by high cholesterol in rabbits.35
Our analysis is in agreement with the effect of polyphenols observed in previous in vitro and in vivo studies. EGCG has been shown to inhibit tumor growth in mouse models of breast cancer by preventing hypoxia-inducible factor-1α and nuclear factor-kappa B activation.36 To the best of our knowledge, no consensus has been derived on the influence of flavonoids on HMGR activity. Recently, it was shown that chokeberry extract, which is rich in polyphenols, inhibited HMGR expression in Caco-2 cells in a dose-dependent manner.37
Previous studies also reported that integration of herbal medicines may provide a more effective and safe means of alleviating lipid disorders.38 A phytoconstituent, n-Octadecanyl-O-α-d-glucopyranosyl(6′→1″)-O-α-d-glucopyranoside isolated from the methanolic extract of Ficus virens, has recently been shown to inhibit HMGR activity.39 The docking analysis of this compound suggested that it does not occupy the catalytic domain, and so is not competitive in binding.40 We observed that the flavonoids have a rigid structure that prevents them from entering the enzyme-active site of HMGR and inhibiting HMG-CoA binding. The phenolic rings of flavonoids can best fit in the NADP(H) binding site (Figure 2). Therefore, they can competitively inhibit NADP(H) binding and may offer some steric hindrance to HMG-CoA binding. The -gallate group of EGCG and the phenol ring of curcumin and THC, which is connected by flexible alkyl chains, can bind to the HMG-CoA binding site. Therefore, EGCG, curcumin, and THC are polyphenols with the greatest potential to inhibit cholesterol biosynthesis.
Conclusion
The findings of the present docking study demonstrate that all the polyphenols tested in this study can bind in the hydrophobic region of the L-domain and cis-loop, leading to the substrate binding cavity, and can sterically hinder HMG-CoA binding. From the study results, it is evident that most polyphenolic rings may pose steric hindrance on NADP+ binding. Among all the tested polyphenols, curcumin, THC, EGCG, and kaempferol can occupy the HMG-CoA binding site and act as competitive inhibitors of substrate binding on the enzyme. The present docking results show that EGCG, curcumin, and THC have strong potential to bind with high affinity. The study results provide a mechanistic rationale for the cholesterol-lowering effect of EGCG, curcumin, and THC.
Gaining a mechanistic insight into the effects of polyphenols on HMGR, a key rate-limiting enzyme, could be useful in drug discovery and development on a pharmacological basis. Moreover, due to the importance of mevalonate pathways in the regulation of cholesterol metabolism, further studies should be carried out to elucidate the in vivo mechanisms of polyphenols that regulate plasma cholesterol through a cascade mechanism mediated by the inactivation of HMGR.
Acknowledgments
There are no patents or products in development or marketed products to declare. This study was supported by grants from the College of Medicine & Health Sciences, United Arab Emirates University, United Arab Emirates. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the paper. The authors would like to thank Professor Keith Bagnall, Department of Anatomy, College of Medicine and Health Sciences, United Arab Emirates University, United Arab Emirates, for critically reading the paper and for providing language editing.
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization [webpage on the Internet]. World health statistics. Geneva, Switzerland: World Health Organization; 2009. Available from: http://www.who.int/gho/publications/world_health_statistics/EN_WHS09_Full.pdf. Accessed: November 15, 2014. | ||
Singh P, Saxena R, Srinivas G, Pande G, Chattopadhyay A. Cholesterol biosynthesis and homeostasis in regulation of the cell cycle. PLoS One. 2013;8(3):e58833. | ||
Stamler J, Daviglus ML, Garside DB, Dyer AR, Greenland P, Neaton JD. Relationship of baseline serum cholesterol levels in 3 large cohorts of younger men to long-term coronary, cardiovascular, and all-cause mortality and to longevity. JAMA. 2000;284(3):311–318. | ||
Sugiyama D, Okamura T, Wantanabe M, et al; NIPPON DATA 80/90 Research Group. Risk of hypercholesterolemia for cardiovascular disease and the population attributable fraction in a 24-year Japanese cohort study. J Atheroscler Thromb. 2015;22(1):95–107. | ||
Carmena R, Duriez P, Fruchart JC. Atherogenic lipoprotein particles in atherosclerosis. Circulation. 2004;109(23 Suppl 1):III2–III7. | ||
Luskey KL, Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J Biol Chem. 1985;260(18):10271–10277. | ||
Endo A. The discovery and development of HMG-CoA reductase inhibitors. J Lipid Res. 1992;33(11):1569–1582. | ||
Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292(5519):1160–1164. | ||
Marcum ZA, Vande Griend JP, Linnebur SA. FDA drug safety communications: a narrative review and clinical considerations for older adults. Am J Geriatr Pharmacother. 2012;10(4):264–271. | ||
de Ferranti S, Ludwig DS. Storm over statins – the controversy surrounding pharmacologic treatment of children. N Engl J Med. 2008;359(13):1309–1312. | ||
Noratto G, Martino HS, Simbo S, Byrne D, Mertens-Talcott SU. Consumption of polyphenol-rich peach and plum juice prevents risk factors for obesity-related metabolic disorders and cardiovascular disease in Zucker rats. J Nutr Biochem. 2015;26(6):633–641. | ||
Yamagata K, Tagami M, Yamori Y. Dietary polyphenols regulate endothelial function and prevent cardiovascular disease. Nutrition. 2015;31(1):28–37. | ||
Zern TL, Wood RJ, Greene C, et al. Grape polyphenols exert a cardioprotective effect in pre- and postmenopausal women by lowering plasma lipids and reducing oxidative stress. J Nutr. 2005;135(8):1911–1917. | ||
Chen ZY, Jiao R, Ma KY. Cholesterol-lowering nutraceuticals and functional foods. J Agric Food Chem. 2008;56(19):8761–8773. | ||
Sesso HD, Gaziano JM, Liu S, Buring JE. Flavonoid intake and the risk of cardiovascular disease in women. Am J Clin Nutr. 2003;77(6):1400–1408. | ||
Cuccioloni M, Mozzicafreddo M, Spina M, et al. Epigallocatechin-3-gallate potently inhibits the in vitro activity of hydroxy-3-methyl-glutaryl-CoA reductase. J Lipid Res. 2011;52(5):897–907. | ||
Zingg JM, Hasan ST, Meydani M. Molecular mechanisms of hypolipidemic effects of curcumin. Biofactors. 2011;39(1):101–121. | ||
Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15(5):411–428. | ||
Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. | ||
Schüttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355. Available from : http://davapc1.bioch.dundee.ac.uk/prodrg/”The PRODRG Server - Daan van Aalten Laboratory. Accessed: October 15, 2014. | ||
Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19(5):819–830. | ||
Lee KH, Park E, Lee HJ, et al. Effects of daily quercetin-rich supplementation on cardiometabolic risks in male smokers. Nutr Res Pract. 2011;5(1):28–33. | ||
Hooper L, Kroon PA, Rimm EB, et al. Flavonoids, flavonoid-rich foods, and cardiovascular risk: a meta-analysis of randomized controlled trials. Am J Clin Nutr. 2008;88(1):38–50. | ||
Davies MJ, Judd JT, Baer DJ, et al. Black tea consumption reduces total and LDL cholesterol in mildly hypercholesterolemic adults. J Nutr. 2003;133(10):3298S–3302S. | ||
Ge H, Liu J, Zhao W, et al. Mechanistic studies for tri-targeted inhibition of enzymes involved in cholesterol biosynthesis by green tea polyphenols. Org Biomol Chem. 2014;12(27):4941–4951. | ||
Wu Q, Li JZ, Zhao TY, et al. Epigallocatechin-3-gallate (EGCG) inhibits 3-hydroxy-3-methylglutaryl-CoA reductase in the presence of glycerol. Pak J Pharm Sci. 2014;27(6):1905–1910. | ||
Ejaz A, Wu D, Kwan P, Meydani M. Curcumin inhibits adipogenesis in 3T3-L1 adipocytes and angiogenesis and obesity in C57/BL mice. J Nutr. 2009;139(5):919–925. | ||
Shin SK, Ha TY, McGregor RA, Choi MS. Long-term curcumin administration protects against atherosclerosis via hepatic regulation of lipoprotein cholesterol metabolism. Mol Nutr Food Res. 2011;55(12):1829–1840. | ||
Pari L, Murugan P. Antihyperlipidemic effect of curcumin and tetrahydrocurcumin in experimental type 2 diabetic rats. Ren Fail. 2007;29(7):881–889. | ||
Karthikesan K, Pari L, Menon VP. Antihyperlipidemic effect of chlorogenic acid and tetrahydrocurcumin in rats subjected to diabetogenic agents. Chem Biol Interact. 2010;188(3):643–650. | ||
Ou JL, Mizushina Y, Wang SY, Chuang DY, Nadar M, Hsu WL. Structure-activity relationship analysis of curcumin analogues on anti-influenza virus activity. FEBS J. 2013;280(22):5829–5840. | ||
Aggarwal BB, Deb L, Prasad S. Curcumin differs from tetrahydrocurcumin for molecular targets, signaling pathways and cellular responses. Molecules. 2014;20(1):185–205. | ||
Lin J, Rexrode KM, Hu F, et al. Dietary intakes of flavonols and flavones and coronary heart disease in US women. Am J Epidemiol. 2007;165(11):1305–1313. | ||
Gebhardt R. Variable influence of kaempferol and myricetin on in vitro hepatocellular cholesterol biosynthesis. Planta Med. 2003;69(12):1071–1074. | ||
Kong L, Luo C, Li X, Zhou Y, He H. The anti-inflammatory effect of kaempferol on early atherosclerosis in high cholesterol fed rabbits. Lipids Health Dis. 2013;12:115. | ||
Gu JW, Makey KL, Tucker KB, et al. EGCG, a major green tea catechin suppresses breast tumor angiogenesis and growth via inhibiting the activation of HIF-1α and NFκB, and VEGF expression. Vasc Cell. 2013;5(1):9. | ||
Kim B, Park Y, Wegner CJ, Bolling BW, Lee J. Polyphenol-rich black chokeberry (Aronia melanocarpa) extract regulates the expression of genes critical for intestinal cholesterol flux in Caco-2 cells. J Nutr Biochem. 2013;24(9):1564–1570. | ||
Zhu PL, Pan SY, Zhou SF, et al. Effects of combined dietary supplementation with fenofibrate and Schisandrae Fructus pulp on lipid and glucose levels and liver function in normal and hypercholesterolemic mice. Drug Des Devel Ther. 2015;9:923–935. | ||
Peschel D, Koerting R, Nass N. Curcumin induces changes in expression of genes involved in cholesterol homeostasis. J Nutr Biochem. 2007;18(2):113–119. | ||
Iqbal D, Khan MS, Khan MS, Ahmad S, Hussain MS, Ali M. Bioactivity guided fractionation and hypolipidemic property of a novel HMG-CoA reductase inhibitor from Ficus virens Ait. Lipids Health Dis. 2015;14:15. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.