Back to Journals » Drug Design, Development and Therapy » Volume 9
Inhibition of HIV-1 by curcumin A, a novel curcumin analog
Authors Kumari N, Kulkarni A, Lin X, McLean C, Ammosova T, Ivanov A, Hipolito M, Nekhai S, Nwulia E
Received 14 April 2015
Accepted for publication 6 June 2015
Published 3 September 2015 Volume 2015:9 Pages 5051—5060
DOI https://doi.org/10.2147/DDDT.S86558
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Shu-Feng Zhou
Namita Kumari,1,2,* Amol A Kulkarni,3,* Xionghao Lin,2 Charlee McLean,1 Tatiana Ammosova,2 Andrey Ivanov,2 Maria Hipolito,1 Sergei Nekhai,2 Evaristus Nwulia1
1Translational Neuroscience Laboratory, 2Department of Medicine, Center for Sickle Cell Disease, College of Medicine, 3College of Pharmacy, Howard University, Washington, DC, USA
*These authors contributed equally to this study
Abstract: Despite the remarkable success of combination antiretroviral therapy at curtailing HIV progression, emergence of drug-resistant viruses, chronic low-grade inflammation, and adverse effects of combination antiretroviral therapy treatments, including metabolic disorders collectively present the impetus for development of newer and safer antiretroviral drugs. Curcumin, a phytochemical compound, was previously reported to have some in vitro anti-HIV and anti-inflammatory activities, but poor bioavailability has limited its clinical utility. To circumvent the bioavailability problem, we derivatized curcumin to sustain retro-aldol decomposition at physiological pH. The lead compound derived, curcumin A, showed increased stability, especially in murine serum where it was stable for up to 25 hours, as compared to curcumin that only had a half-life of 10 hours. Both curcumin and curcumin A showed similar inhibition of one round of HIV-1 infection in cultured lymphoblastoid (also called CEM) T cells (IC50=0.7 µM). But in primary peripheral blood mononuclear cells, curcumin A inhibited HIV-1 more potently (IC50=2 µM) compared to curcumin (IC50=12 µM). Analysis of specific steps of HIV-1 replication showed that curcumin A inhibited HIV-1 reverse transcription, but had no effect on HIV-1 long terminal repeat basal or Tat-induced transcription, or NF-κB-driven transcription at low concentrations that affected reverse transcription. Finally, we showed curcumin A induced expression of HO-1 and decreased cell cycle progression of T cells. Our findings thus indicate that altering the core structure of curcumin could yield more stable compounds with potent antiretroviral and anti-inflammatory activities.
Keywords: curcumin A, HIV-1, reverse transcription, heme oxygenase-1
Introduction
Curcumin, a natural phytochemical derived from turmeric has been reported to have a number of biological effects including inhibition of HIV-1 infection.1,2 Curcumin was shown to inhibit HIV-1 transcription,3 particularly through Tat-activated HIV-1 transcription.3–5 It was also shown to prevent the release of HIV-1 from chronically infected cells,6 inhibit in vitro HIV-1 protease7 and HIV-1 integrase,8,9 and demonstrated in silico binding to HIV-1 reverse transcriptase,10 protease,11 and integrase.12 However, these in vitro effects are yet to be confirmed with live virus in cultured cells. Additionally, curcumin was shown to protect neurons13 and to have a positive effect on spatial memory in HIV-1-associated dementia in a rat model.14
Curcumin affects cell cycle15 and also induces HO-1.2 HO-1 plays a critical role in the regulation of inflammation and immune response.16–18 Induction of HO-1 inhibits HIV-1,19 hepatitis C virus,20 and Ebola virus.21 We recently showed that HO-1 induction led to the production of macrophage inflammatory cytokines and decreased surface expression of CCR-5 in lipopolysaccharide (LPS)-treated monocyte-derived macrophages providing an additional molecular mechanism of HIV-1 inhibition by LPS in macrophages.22 Thus, it was of interest to analyze the effect of curcuminoids on HO-1 expression.
Despite many reported pharmacological properties of curcumin23–25 and its low toxicity in clinical trials,2,26 it demonstrated poor absorption, rapid metabolism, rapid systemic elimination,27–29 and overall poor bioavailability.28–31 We sought to improve the bioavailability of curcumin by developing structural analogs of curcumin by taking a previously reported approach in which β-diketone moiety of curcumin was removed,28 and to test these analogs for HIV-1 inhibition and HO-1 induction.
We synthesized a novel curcuminoid, curcumin A and obtained a pure compound. We analyzed stability of curcumin A in parallel to curcumin in mouse serum, media, fetal bovine serum-free media, and complete media. We also analyzed stability of curcumin and curcumin A in phosphate-buffered saline (PBS) and in PBS supplemented with 10% bovine serum albumin (BSA). We then tested curcumin A and the parental curcumin for HIV-1 inhibition in cultured T cells and primary peripheral blood mononuclear cells (PBMCs) infected with vesicular stomatitis virus glycorotein-pseudotyped HIV-1. We also determined toxicity of the compounds in T cells and PBMCs. To establish a mechanism of the curcumin-mediated inhibition of HIV-1, we analyzed the effect of curcumin on early and later stages of HIV-1 infection using single round HIV-1 infection with vesicular stomatitis virus glycorotein-pseudotyped HIV-1. Finally, we tested HO-1 induction by curcumin and curcumin A and their effect on cell cycle progression. Our study demonstrates that curcumin A is a stable curcuminoid that has strong antiretroviral activity by targeting HIV-1 reverse transcription (RT) and inducing HO-1. Thus, altering the core structure of curcumin could yield more stable compounds with potent antiretroviral and anti-inflammatory activities.
Materials and methods
Cells were obtained from ATCC, (American Type Culture Collection, Manassas, VA, USA). PBMCs were purchased from Astarte Biologics (Redmond, WA, USA).
Plasmids
HIV-1 proviral vector pNL4-3.Luc.R-E- (courtesy of Prof Nathaniel Landau, NYU School of Medicine, New York, NY, USA) was obtained from the NIH AIDS Research and Reference Reagent Program. HIV-1 LTR luciferase expression vectors were kindly provided by Dr Manuel López-Cabrera (Unidad de Biología Molecular, Madrid, Spain):32 HIV-1 LTR (−105 to +77) followed by the luciferase reporter gene, (HIV LTR 2× NF-κB 3× SP1), HIV-1 LTR, NF-κB and SP1 sites followed by luciferase reporter gene.
Synthesis of curcumin A
The chemical synthesis of the acyclic analog of curcumin (curcumin A) was accomplished via a slight modification of the previously reported procedure.28 In brief, acetic acid (50 mL) was saturated with hydrogen chloride for a period of 1 hour at 0°C. A mixture of vanillin (3.04 g, 20 mmol, 2.0 equiv) and acetone (580 mg, 10 mmol, 1.0 equiv) was slowly added to acetic acid. The reaction mixture was slowly brought to room temperature and stirred for 48 hours. The crude reaction mixture was poured into ice-cold water (200 mL) and the precipitated solid was isolated via vacuum filtration. It was dried and purified using silica gel flash column chromatography. Gradient elution with 30%–60% ethyl acetate in hexanes resulted in the isolation of the desired product (1.93 g, 59% yield) as yellowish orange solid.
Solubility of curcumin A
To explore the solubility of curcumin A, an excess amount of curcumin A was added to 1 mL of polyethylene glycol 400 in a 5 mL stopper vial, vortex mixed, equilibrated, and analyzed by high-performance liquid chromatography, using a previously established technique.33 The solubility was 86.17±4.34 mg/mL, suggesting that curcumin A has good solubility in a pharmaceutically approved excipient.
Single round HIV-1 replication assay
CEM-T cells or PBMCs were infected with VSVG-pseudotyped pNL4-3.Luc.R-E-virus (HIV-1 Luc) prepared as previously described.34 PBMCs were stimulated by treating with 2.5 μg/mL phytohemagglutinin (PHA) for 24 hours and then activated for another 24 hours with 10 units/mL IL-2 before the infection with HIV-1 Luc. PBMCs were infected with HIV-1 Luc, cultured at 0.5×106 cells/mL in 6-well plates at 37°C and 5% CO2 for 24 hours and then treated with indicated concentration of compounds. The cells were collected after 48 hours in culture, washed with PBS, and resuspended in 100 μL of PBS. Then, 100 μL of reconstituted luciferase buffer (Luclite Kit; PerkinElmer Inc., Waltham, MA, USA) was added to each sample and, after 10 minutes incubation, the lysates were transferred into white plates (PerkinElmer Inc.) and luminescence measured using Labsystems Luminoscan RT equipment (PerkinElmer Inc.).
Calcein-AM uptake cell viability assays
PBMCs were cultured as described above in 96-well plates at 37°C. To measure cytotoxicity with calcein, cells were supplemented with 0.2 μM calcein-AM (Thermo Fisher Scientific, Waltham, MA, USA) for 30 minutes at 37°C. Fluorescence was measured using the luminescence spectrometer implementing an excitation wavelength of 495 nm and emission filters at 515 nm. To measure cellular viability with trypan blue, the cells were supplemented with 0.2% trypan blue, transferred to a plastic disposable counting chamber, and counted on a Cellometer Automatic Cell Counter (Nexcelcom Bioscience, Lawrence, MA, USA).
Cell cycle analysis of CEM-T cells treated with curcumin A
Approximately one million cells were fixed in 70% ethanol at −20°C for 2 hours and stained with propidium iodide (10 mg/mL) containing RNAse A (1 mg/mL) for 30 minutes. The data were acquired in BD FACS Calibur (BD Biosciences, San Jose, CA, USA), and analyzed using FlowJo software. Unpaired Student’s t-test was used to determine statistical significance.
Analysis of HO-1 messenger (m)RNA expression
CEM-T cells or PBMCs were treated with curcumin or curcumin A (2 μM) for 24 hours. Total RNA was extracted from cultured cells using TRIzol reagent according to the manufacturer’s protocol (Thermo Fisher Scientific). Total RNA (100 ng) was reverse-transcribed to complementary (c)DNA using Superscript™ RT-PCR (reverse transcription polymerase chain reaction) kit (Thermo Fisher Scientific); hexamers and oligo-dT were used as primers. For RT-PCR analysis, cDNA was amplified using Roche LightCycler 480 (Hoffman-La Roche Ltd., Basel, Switzerland) and SYBR Green1 Master mix (Hoffman-La Roche Ltd.). PCR was carried with denaturation at 95°C for 10 seconds, annealing at 60°C for 10 seconds, and extension at 72°C for 10 seconds for 45 cycles. For quantification of mRNA levels for HO-1, 18S ribosomal (r)RNA was used as a house keeping normalization standard. The following primer sequences were used for HO-1, forward-CTTCTTCACCTTCCCCAACA; reverse-GCTCTGGTCCTTGGTGTCAT, amplicon size 193; for 18S rRNA, forward-GCTGTTGCTACATCGACCTTT; reverse-CTCCAGGTTTTGCAACCAGT, amplicon size 168. Mean crossing point values for HO-1 and 18S rRNA were determined and relative changes in gene expression were analyzed using an algorithm known as ΔΔCt method. Unpaired Student’s t-test was used to determine statistical significance.
Quantitative PCR for viral DNA
CEM-T cells or stimulated PBMCs were infected with HIV-1 Luc and treated with 2 μM curcumin, curcumin A or 1 μM azidothymidine (AZT) and further cultured for 48 hours. Total DNA was extracted from 4×106 cells using lysis buffer (10 mM Tris-HCl pH 8, 10 mM ethylenediaminetetraacetic acid, 5 mM NaCl, 200 μg/mL proteinase K). The cells were lysed for 20–30 minutes at room temperature and proteinase K was inactivated by heating to 95°C for 5 minutes. For the RT-PCR analysis, 100 ng DNA was amplified using Roche Light Cycler 480 and SYBR Green1 Master mix. PCR was carried out with initial preincubation for 5 minutes at 45°C and then for 3 minutes at 95°C followed by 45 cycles of denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 45 seconds, and final extension at 72°C for 10 seconds. Quantification of early-LTR and late-LTR was carried out using β-globin DNA as a normalization standard. Primer sequences for early-LTR, forward-GGCTAACTAGGGAACCCACTG, reverse-CTGCTAGAGATTTTCCACACTGAC; for late-LTR forward-TGTGTGCCCGTCTGTTGTGT, reverse-GAGTCCTGCGTCGAGAGATC; and for globin, forward-CAACCTCAAACAGACACCATGG, reverse-TCCACGTTCACCTTGCCC.35 Mean crossing point values for early-LTR, late-LTR, and β-globin were determined and ΔΔCt method was used to calculate relative expression levels. Unpaired Student’s t-test was used to test statistical significance.
Analysis of curcumin A stability
Serum incubation
Curcumin or curcumin A was dissolved in dimethylsulfoxide (DMSO) (10 mM), and then diluted with PBS to 100 μM, which was mixed with an equal volume of mouse serum. The final concentration of curcumin or curcumin A was 50 μM, and the amount of organic solvent in the mixture was less than 1% (v/v). The mixture was incubated at 37°C and samples were collected at 0 hours, 4 hours, 8 hours, 12 hours, 16 hours, 20 hours, and 24 hours. The samples (60 μL) were mixed with 240 μL of cold acetone, vortexed for 2 minutes, kept at −20°C for 30 minutes, and then proteins were removed by centrifugation at 13,000× g for 5 minutes. The supernatant was transferred to a clean test tube and evaporated to dryness using a Speedvac concentrator. The pellet was reconstituted in 50 μL of acetonitrile, and a 10 μL aliquot was injected for analysis.
Media or PBS incubation
Curcumin or curcumin A was dissolved in DMSO (10 mM), and then diluted with Roswell Park Memorial Institute (RPMI) 1640 media, PBS or PBS supplemented with 10% BSA. The solutions were incubated at 37°C and samples were collected at different time points within 24 hours incubation. The sample (100 μL) was mixed with 400 μL of cold acetone, vortexed for 2 minutes, kept at −20°C for 30 minutes, and then proteins were removed by centrifugation at 13,000× g for 5 minutes. The supernatant was transferred to a clean test tube and evaporated to dryness using a Speedvac concentrator. The pellet was reconstituted in 50 μL of acetonitrile, and a 10 μL aliquot was injected onto the nano C18 column for the liquid chromatography-mass spectrometry (LC-MS) for analysis.
Nano-LC-MS condition
The samples were loaded onto a nano-C18 column attached to Shimadzu nano-LC coupled in-line to LTQ Orbitrap XL tandem mass spectrometer (Thermo Fisher Scientific). The injection volume was 10 μL. The mobile phase consisted of a 0.1% formic acid aqueous solution (A) and a 0.1% formic acid acetonitrile solution (B). The gradient elution program was as follows: 0–6.02 minutes, 1% B; 6.02–6.11 minutes, 1%–2% B; 6.11–20 minutes, 2%–80% B; 20–25 minutes, 80% B; 25–30 minutes, 80%–85% B; 30–31 minutes, 80%–2% B; 31–40 minutes, 2% B (v/v). The flow rate was set to 600 nL/min. The compounds were ionized by electrospray ionization and detected by Orbitrap at 30,000 mass resolution (full scan, m/z 150-2,000). The spray voltage, capillary temperature, and capillary voltage were set to 2.0 kV, 200°C, and 39.5 V, respectively.
Results and discussion
Synthesis of curcumin A
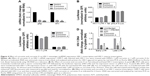
Presence of β-diketone moiety in curcumin makes it prone to decomposition at physiological pH (see structure in Figure 1A). To exclude the possibility that curcumin contained contaminants, we analyzed its composition by LC-MS analysis and did not find demethoxycurcumin or bisdemethoxycurcumin, typical contaminants (Figure 1A). To improve curcumin stability, we synthesized curcumin A (as described in Materials and methods) in which β-diketone moiety was removed (Figure 1B). Removal of the β-diketone moiety from curcumin yielded yellowish orange solid of molecular weight 327.1232 Mp 100–102°C.
Curcumin A structure was verified by nuclear magnetic resonance analysis (1H-NMR) (CDCl3, 400 MHz, ppm) δ 7.66 (d, J =15.6 Hz, 2H), 7.18 (dd, J =8.0, 1.6 Hz, 2H), 7.11 (d, J =2 Hz, 2H), 6.96 (d, J =1.6 Hz, 2H), 6.91 (d, J =9.6 Hz, 2H), 5.96 (s, 2H), 3.95 (s, 6H) (Figure 1C). Other spectroscopic data (carbon-13 nuclear magnetic resonance spectroscopy, infrared spectroscopy, and high-resolution mass spectroscopy) for curcumin A were consistent with the previously reported literature and its predicted chemical structure (not shown). Curcumin A purity was further analyzed by LC-MS high resolution mass spectrometry which shows a single peak with the expected molecular weight (Figure 1D). Thus we obtained pure curcumin A with the expected structure which was analyzed for its stability and inhibition of HIV-1.
Curcumin A has an improved stability
To analyze stability of curcumin A and compare it to curcumin, the compounds were incubated with mouse plasma, in complete or minimal cell culture media, and in PBS. The compounds were extracted from the incubation mixtures and resolved by nano-LC-MS mass spectrometry as described in Materials and methods. While curcumin had a half-life of 10 hours in mouse serum, curcumin A was stable for up to 24 hours of incubation (Figure 2A). In serum-free media, both curcumin and curcumin A were degraded, but curcumin A had a longer half-life (about 3 hours) compared to curcumin (about 1 hour) (Figure 2B). In complete media, curcumin and curcumin A were equally and relatively stable (Figure 2C). Incubation in PBS again showed increased stability of curcumin A (Figure 2D). To test whether BSA has a stabilizing effect on curcumin, curcumin and curcumin A were incubated in PBS supplemented with 10% BSA (Figure 2E). No stabilization and even a slight increase in degradation of curcumin was observed suggesting that BSA has no stabilizing effect on curcumin (Figure 2E). Taken together, curcumin A has improved stability and longer half-life, in serum, serum-free media, and PBS.
Curcumin A inhibits one round of HIV-1 infection
The effects of curcumin and curcumin A on one round of HIV-1 infection were first analyzed in cultured CEM-T cells infected with VSVG-pseudotyped HIV-1 pNL4-3 virus expressing luciferase in place of nef (HIV-1 Luc).34 The cells were treated with the compounds for 24 hours and luciferase activity was measured as an indicator of HIV-1 replication. One round HIV-1 infection was equally inhibited by curcumin (the half maximal inhibitory concentration [IC50] =0.7 μM) and curcumin A (IC50=0.8 μM) (Figure 3A). Viability of CEM-T cells was analyzed using calcein-AM assay which measured fluorescence of the calcein converted intracellularly from the non-fluorescent calcein-AM taken up by the cells. Curcumin and curcumin A reduced viability of CEM-T cells when added to the cells for 24 hours, with curcumin being slightly more toxic (IC50=1.26 μM) than curcumin A (IC50=2.4 μM) (Figure 3B). We next analyzed the effect of curcumin and curcumin A on single round HIV-1 infection in primary PBMCs. PBMCs were activated by treatment with PHA and IL-2 (see Materials and methods for details), infected with HIV-1 Luc for 24 hours and then treated with curcumin or curcumin A for another 24 hours. Both curcumin and curcumin A markedly inhibited HIV-1 infection with curcumin A being more potent (IC50=2 μM) than curcumin (IC50=12 μM) (Figure 3C). Both curcumin and curcumin A reduced viability of PBMCs as analyzed using trypan blue, with IC50s of 35 μM and 22 μM, respectively (Figure 3D). Thus, curcumin A inhibits one round HIV-1 infection comparable to curcumin in CEM-T cells and displays more potent activity in primary PBMCs. Also, curcumin A demonstrated a better therapeutic window in both cultured CEM-T cells and PBMCs.
Curcumin and curcumin A inhibit HIV-1 RT
Previously, curcumin was shown to inhibit HIV-1 transcription by having an effect on chromatin remodeling.36 To elucidate if curcumin and curcumin A inhibited HIV-1 transcription in the context of single round HIV-1 infection, we analyzed the expression of HIV-1 encoded gag and env mRNAs in CEM-T cells infected with HIV-1 Luc virus. Expression of these mRNAs was significantly reduced in infected cells treated with curcumin or curcumin A (Figure 4A). To determine whether the inhibition was due to the effect of curcumin or curcumin A on HIV-1 transcription, 293T cells were transfected with a combination of a reporter expressing luciferase under the control of HIV-1 LTR and a Tat expressing vector or proviral pNL4-3 vector expressing luciferase in place of nef. Neither curcumin nor curcumin A at 2 μM concentrations had an effect on Tat-induced HIV-1 transcription or transcription from proviral pNL4-3 plasmid (Figure 4B). We also analyzed the effect of curcumin and curcumin A on basal HIV-1 LTR transcription and NF-kB driven transcription in 293T cells transfected with HIV-1 LTR luciferase vector or HIV LTR 2× NF-κB 3× SP1 vector (described in Materials and methods). There was no effect of curcumin or curcumin A on basal HIV-1 transcription or NF-kB driven transcription (Figure 4C) at 2 μM concentration. We reasoned that curcumin and curcumin A might affect an earlier stage of HIV-1 infection and thus indirectly reduce the subsequent HIV-1 transcription step. We analyzed early and late RT by quantifying HIV-1 DNA for early and late LTR.35 Both curcumin and curcumin A inhibited early LTR similarly or better than the established HIV-1 inhibitor, AZT (Figure 4D). In contrast, only curcumin A inhibited late LTR formation, similar to AZT, whereas curcumin had no significant effect (Figure 4D). Thus, curcumin inhibited one round HIV-1 infection with an inhibitory effect on HIV-1 RT, while curcumin A showed more potent inhibition of both early and late HIV-1 RT.
Curcumin and curcumin A induce expression of HO-1
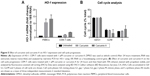
Previously, curcumin was shown to induce HO-1.2 Activation of HO-1 by heme was also shown to inhibit HIV-1.19 Thus we examined the effects of curcumin and curcumin A on HO-1 in primary PBMCs and also in cultured CEM-T cells. The cells were treated with 1 μM curcumin A or curcumin and HO-1 expression was analyzed by mRNA quantification with quantitative PCR using 18S RNA for normalization. Both curcumin A and curcumin induced HO-1 in CEM-T cells and PBMCs (Figure 5A). Thus, the HIV-1 inhibitory effects of curcumin and curcumin A could partly be due to the activation of HO-1.
Treatment with curcumin A affects cell cycle progression
Curcumin and its analogs were previously shown to have an effect on cell cycle progression and to cause apoptosis of cancer cells.37 Thus we examined the effects of curcumin A and curcumin on the cell cycle progression of CEM-T cells. While curcumin had no effect on the cell cycle progression of CEM-T cells, the cells treated with curcumin A showed increased S phase accumulation (1.4 times) and decreased G2/M phase accumulation (three times) (Figure 5B). Thus this reduction of the cell cycle progression could be contributory to the effect of curcumin A.
Conclusion
We show here that curcumin A, which we derivatized from curcumin, has improved stability in serum and serum-free media over the parent compound, and also retained the ability of the parent compound to inhibit HIV-1 infection in CEM-T cells and PBMCs. Phase 1 clinical trials of curcumin have shown that efficacy of curcumin is limited by its poor bioavailability, resultant from its poor absorption, rapid metabolism, and rapid systemic elimination.27 This underscores the relevance of development of derivatives which retain the rich biological activities of curcumin, but also withstand the rapid degradation that has plagued the desired pharmacodynamics effect of the parent compound: curcumin A, the derivative we obtained in pure form, is stable in serum up to 24 hours as opposed to curcumin which is degraded by 50% within 10 hours. We also found that at 2 μM concentration curcumin A and curcumin inhibited HIV-1 RT while having no effect on HIV-1 transcription. Further analysis of the mechanism of HIV-1 inhibition pointed to HO-1 induction, in line with earlier reports of HIV-1 inhibition by HO-1,19 and alteration of cell cycle kinetics as putative mechanisms. The intended route of future administration of curcumin A is intranasal. Recently, curcumin was formulated as a nanoemulsion and successfully tested for intranasal delivery.33 We have also developed micro- and nano-emulsions of curcumin A and are now optimizing these proprietary formulations for brain delivery through intranasal administration. In conclusion, the improved stability of curcumin A over curcumin, and the retention of the in vitro anti-HIV-1 property of curcumin hold promise for the future potential clinical benefit of this derivative in antiretroviral therapy.
Acknowledgments
This project has been funded in part with funds from the: National Institutes of Health NCI contract number HHSN261200800001E (EN); NIH Research Grants 5R01MH091460-03 (EN), 1P50HL118006 (SN), 1R01HL125005 (SN), U19AI109664 (SN) and 5G12MD007597 (SN); Dana Foundations Clinical Neuroscience Award (EN); and District of Columbia Developmental Center for AIDS Research grant (P30AI087714) (AK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
AAK and EN developed curcumin and curcuminoids, including curcumin A. AAK, EN, and SN conceived the study. NK and CM analyzed antiviral and cellular effects of curcumin A. XL and TA conducted MS and stability analysis. AI purified HIV-1 virus and helped to conduct antiviral analysis. MH helped in preparing the manuscript. AAK, SN, and EN analyzed the data and wrote the manuscript. All authors contributed toward data analysis, drafting, and revising the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Itokawa H, Shi Q, Akiyama T, Morris-Natschke SL, Lee KH. Recent advances in the investigation of curcuminoids. Chin Med. 2008;3:11. | ||
Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 2013;15(1):195–218. | ||
D’Amours G, Langlois M, Mathonnet G, et al. SNP arrays: comparing diagnostic yields for four platforms in children with developmental delay. BMC Med Genomics. 2014;7:70. | ||
Issuriya A, Kumarnsit E, Wattanapiromsakul C, Vongvatcharanon U. Histological studies of neuroprotective effects of Curcuma longa Linn. on neuronal loss induced by dexamethasone treatment in the rat hippocampus. Acta Histochem. 2014;116(8):1443–1453. | ||
Guan WJ, Zheng XY, Zheng JP, et al. Small airway dysfunction in asymptomatic bronchial hyperresponsiveness and asthma. Allergy. 2014;69(9):1258–1259. | ||
Barquero AA, Davola ME, Riva DA, Mersich SE, Alche LE. Naturally occurring compounds elicit HIV-1 replication in chronically infected promonocytic cells. Biomed Res Int. 2014;2014:989101. | ||
Jamil-Copley S, Bokan R, Kojodjojo P, et al. Noninvasive electrocardiographic mapping to guide ablation of outflow tract ventricular arrhythmias. Heart Rhythm. 2014;11(4):587–594. | ||
Tanaka R, Tsujii H, Yamada T, et al. Novel 3alpha-methoxyserrat-14-en-21beta-ol (PJ-1) and 3beta-methoxyserrat-14-en-21beta-ol (PJ-2)-curcumin, kojic acid, quercetin, and baicalein conjugates as HIV agents. Bioorg Med Chem. 2009;17(14):5238–5246. | ||
S P. A second case with arrhythmogenic cardiomyopathy, provocable Brugada ECG and SCN5A mutation. Int J Cardiol. 2014;171(3):e117–e118. | ||
Seal A, Aykkal R, Babu RO, Ghosh M. Docking study of HIV-1 reverse transcriptase with phytochemicals. Bioinformation. 2011;5(10):430–439. | ||
Vajragupta O, Boonchoong P, Morris GM, Olson AJ. Active site binding modes of curcumin in HIV-1 protease and integrase. Bioorg Med Chem Lett. 2005;15(14):3364–3368. | ||
Mazumder A, Raghavan K, Weinstein J, Kohn KW, Pommier Y. Inhibition of human immunodeficiency virus type-1 integrase by curcumin. Biochem Pharmacol. 1995;49(8):1165–1170. | ||
Guo L, Xing Y, Pan R, et al. Curcumin protects microglia and primary rat cortical neurons against HIV-1 gp120-mediated inflammation and apoptosis. PloS One. 2013;8(8):e70565. | ||
Tang H, Lu D, Pan R, Qin X, Xiong H, Dong J. Curcumin improves spatial memory impairment induced by human immunodeficiency virus type 1 glycoprotein 120 V3 loop peptide in rats. Life Sci. 2009;85(1–2):1–10. | ||
Shehzad A, Lee J, Lee YS. Curcumin in various cancers. Biofactors. 2013;39(1):56–68. | ||
Takaki S, Takeyama N, Kajita Y, et al. Beneficial effects of the heme oxygenase-1/carbon monoxide system in patients with severe sepsis/septic shock. Intensive Care Med. 2010;36(1):42–48. | ||
Slebos DJ, Ryter SW, Choi AM. Heme oxygenase-1 and carbon monoxide in pulmonary medicine. Respir Res. 2003;4:7. | ||
Roach JP, Moore EE, Partrick DA, et al. Heme oxygenase-1 induction in macrophages by a hemoglobin-based oxygen carrier reduces endotoxin-stimulated cytokine secretion. Shock. 2009;31(3):251–257. | ||
Devadas K, Hewlett IK, Dhawan S. Lipopolysaccharide suppresses HIV-1 replication in human monocytes by protein kinase C-dependent heme oxygenase-1 induction. J Leukoc Biol. 2010;87(5):915–924. | ||
Chen MH, Lee MY, Chuang JJ, et al. Curcumin inhibits HCV replication by induction of heme oxygenase-1 and suppression of AKT. Int J Mol Med. 2012;30(5):1021–1028. | ||
Hill-Batorski L, Halfmann P, Neumann G, Kawaoka Y. The cytoprotective enzyme heme oxygenase-1 suppresses Ebola virus replication. J Virol. 2013;87(24):13795–13802. | ||
Zhou ZH, Kumari N, Nekhai S, et al. Heme oxygenase-1 induction alters chemokine regulation and ameliorates human immunodeficiency virus-type-1 infection in lipopolysaccharide-stimulated macrophages. Biochem Biophys Res Commun. 2013;435(3):373–377. | ||
Bhullar KS, Jha A, Youssef D, Rupasinghe HP. Curcumin and its carbocyclic analogs: structure-activity in relation to antioxidant and selected biological properties. Molecules. 2013;18(5):5389–5404. | ||
S PM, M AB, Tejaswi S, Shetty S, K SH. Effect of power bleaching on the fluorosis stained anterior teeth case series. J Clin Diagn Res. 2014;8(8):ZJ01–ZJ03. | ||
Anbumani S, Arunai Nambi Raj N, S Prabhakar G, et al. Quantification of uncertainties in conventional plan evaluation methods in Intensity Modulated Radiation Therapy. J BUON. 2014;19(1):297–303. | ||
Epstein J, Sanderson IR, Macdonald TT. Curcumin as a therapeutic agent: the evidence from in vitro, animal and human studies. Br J Nutr. 2010;103(11):1545–1557. | ||
Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807–818. | ||
Bisht S, Maitra A. Systemic delivery of curcumin: 21st century solutions for an ancient conundrum. Curr Drug Discov Technol. 2009;6(3):192–199. | ||
Gandapu U, Chaitanya RK, Kishore G, Reddy RC, Kondapi AK. Curcumin-loaded apotransferrin nanoparticles provide efficient cellular uptake and effectively inhibit HIV-1 replication in vitro. PloS One. 2011;6(8):e23388. | ||
S P, S PK, Jb A. Increased serum alkaline phosphatase and serum phosphate as predictors of mortality after stroke. J Clin Diagn Res. 2014;8(8):CC01–CC03. | ||
Parthasarathy SD, Bridges SM, McGrath CP, Au TK, Wong HM, Yiu CK. The Relation Between Caregivers’ Multiliterate Reading Habits and Their Children’s Oral Health Status. Interact J Med Res. 2014;3(3):e13. | ||
Gomez-Gonzalo M, Carretero M, Rullas J, et al. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NF-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J Biol Chem. 2001;276(38):35435–35443. | ||
Sood S, Jain K, Gowthamarajan K. Optimization of curcumin nanoemulsion for intranasal delivery using design of experiment and its toxicity assessment. Colloids Surf B Biointerfaces. 2014;113:330–337. | ||
Debebe Z, Ammosova T, Breuer D, et al. Iron chelators of the di-2-pyridylketone thiosemicarbazone and 2-benzoylpyridine thiosemicarbazone series inhibit HIV-1 transcription: identification of novel cellular targets – iron, cyclin-dependent kinase (CDK) 2, and CDK9. Mol Pharmacol. 2011;79(1):185–196. | ||
Vermeire J, Naessens E, Vanderstraeten H, et al. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PloS One. 2012;7(12):e50859. | ||
Zhang HS, Ruan Z, Sang WW. HDAC1/NFkappaB pathway is involved in curcumin inhibiting of Tat-mediated long terminal repeat transactivation. J Cell Physiol. 2011;226(12):3385–3391. | ||
Lee DS, Lee MK, Kim JH. Curcumin induces cell cycle arrest and apoptosis in human osteosarcoma (HOS) cells. Anticancer Res. 2009;29(12):5039–5044. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.