Back to Journals » Drug Design, Development and Therapy » Volume 13
In silico and in vitro studies of lupeol and iso-orientin as potential antidiabetic agents in a rat model
Authors Malik A , Jamil U, Butt TT, Waquar S , Gan SH , Shafique H, Jafar TH
Received 26 October 2018
Accepted for publication 14 March 2019
Published 6 May 2019 Volume 2019:13 Pages 1501—1513
DOI https://doi.org/10.2147/DDDT.S176698
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Manfred Ogris
Arif Malik,1 Uzma Jamil,2 Tariq Tahir Butt,3 Sulayman Waquar,1 Siew Hua Gan,4 Hassan Shafique,1 Tassadaq Hussain Jafar1
1Institute of Molecular Biology and Biotechnology (IMBB), The University of Lahore, Lahore, Pakistan; 2Department of Physiology, Shalamar Medical and Dental College Lahore, Lahore, Pakistan; 3Department of Biochemistry, Khawaja Muhammad Safdar Medical College, Sialkot, Pakistan; 4School of Pharmacy, Monash University Malaysia, Bandar Sunway, Selangor Darul Ehsan 47500, Malaysia
Background: In silico characterization can help to explain the interaction between molecules and predict three-dimensional structures. Various studies have confirmed the glucose-lowering effects of plant extracts, ie, lupeol and iso-orientin, which enable them to be used as antidiabetic agents.
Purpose: Aims of the present study were to evaluate the hypoglycemic activities of lupeol and iso-orientin in a rat model. The study proposed the effects of alloxan on blood glucose level, body weight, and oxidative stress.
Materials and Methods: Thirty (n=30) Wistar albino rats were divided into six groups and were subjected to different combinations of the compounds. Levels of different stress markers, ie, malondialdehyde, superoxide dismutase, catalase, nitric oxide, glutathione, glutathione peroxide, glutathione reductase, and blood glucose levels were estimated with their respective methods. Whereas, for their in silico analysis, identified target proteins, GPR40, glucose-6-phosphatase, UCP2, glycogen phosphorylase, aldose reductase, and glucose transporter-4 were docked with lupeol and iso-orientin. Three-dimensional structures were predicted by ERRAT, Rampage, Verify3D, threading and homology approaches.
Results: Blood glucose levels were significantly increased in rats receiving intraperitoneal injection of alloxan (208±6.94 mg/dL) as compared to controls (90±7.38 mg/dL). Infected rats were administered plant extracts; combined treatment of both extracts (lupeol+iso-orientin) significantly reduced the levels of blood glucose (129.06±6.29 mg/dL) and improved the antioxidant status. Fifteen structures of each selected protein were evaluated using various techniques. Consequently, satisfactory quality factors [GPR40 (96.41%), glucose-6-phosphatase (96.56%), UCP2 (72.56%), glycogen phosphorylase (87.24%), aldose reductase (82.46%), and glucose transporter-4 (94.29%)] were selected. Molecular docking revealed interacting residues, effective drug properties and their binding affinities (ie, −8.9 to −12.6 Kcal/mol).
Conclusion: Results of the study affirmed the antidiabetic activities of lupeol and iso-orientin. Administration of these extracts (either individually or in combination) significantly reduced blood glucose levels and oxidative stress. Hence, it may be considered beneficial in the treatment of diabetes.
Keywords: AutoDock vina, molecular docking, Iso-orientin, lupeol, diabetes, GLUT-4
Introduction
Lupeol is the bioactive compound present in Cassia fistula. C. fistula belongs to the Caesal piniaceae family commonly known as Amaltas, Indian laburnum, and golden showers. It is widely used in different traditional medicines such as Ayurveda, Unani, and Chinese.1 It grows in East Africa, South Africa, China, Brazil, India, Nepal, Mexico, Mauritius, and Pakistan.2 In Pakistan, it is widely cultivated in the east of the Indus plains continuing north of the Himalayas, it grows to an approximate height of 1,200 m.3 Whereas, iso-orientin is the active compound of Parkinsonia aculeata L (Leguminosae), a small, spiny tree commonly known as Retama, Mexican Palo Verde, Jerusalem thorn, and Jelly bean tree and is generally found in arid or seasonally flooded areas. The tree is widely distributed in tropical regions of America, India, Hawaii, Egypt, Middle East, Italy, Cyprus, tropical Africa, Australia, and Pakistan. In Pakistan, it is abundantly found along grand trunk roads as an avenue.4
Diabetes mellitus is a chronic metabolic disorder that is characterized by an increase in blood glucose levels and altered metabolism of proteins, carbohydrates, and lipids.5 The word “diabetes“ was derived from the Greek language which literally meant ”to siphon or drain off” which indicates urination, whereas the word “mellitus“ is Latin and means “sweet“, which can be further classified into two groups. Acute metabolic complication is usually short-lived and is characterized by hyperosmolar non-ketonic coma, ketoacidosis, and hypoglycemia. While the other type is a late systemic complication commonly known as chronic type that includes diabetic nephropathy, atherosclerosis, infections, diabetic neuropathy, retinopathy, and micro-angiopathy.6 Among all types of diabetes, two are the most abundant forms. Type I diabetes, also known as insulin-dependent diabetes mellitus, which is an autoimmune disorder, and type II diabetes, which is characterized by functional disruption of insulin action due to the disturbance of receptor or transporters on the cell membrane.7
Glucose enters the cells via GLUT-4, present on the cell membrane. Therefore, reduction of GLUT-4 in muscles contributes to insulin resistance, characteristic features seen in type II diabetes, gestational diabetes, and impaired glucose tolerance. Increased intake of glucose initiates polyol pathway comprised of enzymes, ie, aldose reductase (which converts glucose into sorbitol) and glucose-6-phosphatase that mainly originates from the kidneys and liver, and is expressed in the pancreas. Evident from some studies, increased glucose-6-phosphatase or decreased glucokinase is responsible for the endogenous glucose production in type 2 diabetes.8 GPCR40, also recognized as FFA1, is expressed in the pancreatic β-cells and is stimulated by medium and long chains of fatty acids. Activation of GPCR40 in pancreatic β-cells contributes to insulin secretion, which has a significant effect against diabetes. Another member of the mitochondrial inner membrane carrier family, UCP2, is abundantly present in pancreatic β-cells. UCP2 has structural homology with two other subclades (UCP1 and UCP3). It is considered an important part of β-cells that is involved in type II diabetes. Moreover, UCP2 decreases ATP production via mitochondrial proton leakage.9
Various studies have confirmed the anti-hyperglycemic effects of plant extracts and herbal formulations which have potential to be used as antidiabetic tonics. In comparison to synthetic drugs, herbal medicines are more often considered to have less harmful side effects.10 Investigation of P. aculeata indicated that the leaves of the plant which contain C-glycosylflavones vitexin, isovitexin, orientin, and iso-orientin have some antidiabetic activities. In fact, the bark, which contains flavonoids, alkoloids, steroids, and tannis also have antidiabetic effects.11 C. fistula is important in traditional medicines where its dried seeds tend to show marked hypoglycemic activity. The fruits of C. fistula are used for treatment of disorders relating to the chest, throat, liver, and eyes, moreover, it is also beneficial as abortifacient and antipyretic.12 The seed powder of C. fistula is also used to treat amoebiasis. Additionally, the seeds of C. fistula are also used for their emetic (due to its cathartic properties) and antipyretic activities.13
Numerous biological problems have been tackled by utilizing in silico approaches12 which can explain the interacting molecules and predict three-dimensional (3D) structures. In this study, appropriate phytochemicals and 3D structures were predicted by threading and homology modeling approaches. The aim of the current study was to conduct in silicoand in vitro analysis of lupeol and iso-orientin as potential antidiabetic agents in a rat model.
Materials and methods
Sample collection
The experimental samples of plant extracts lupeol and iso-orientin were purchased from Phoenix Chemicals, Lahore, Pakistan. The extracts were then subjected to non-polar medium, they were dissolved in approximately 1,000 mL of ethanol for the extraction of their potential ingredients and these dissolved extracts were then stored at room temperature. They were finally administered to the subjects at a rate of 200 mg/kg body weight.
Experimental animals and experimental design
Adult albino rats (Rattus norvegicus) aged 4–5 months were used as test subjects in the current study. The rats were purchased from the animal house of Institute of Molecular Biology and Biotechnology (IMBB), The University of Lahore, Pakistan. They were acclimatized to the lab environment for at least 1 week prior to the experiment and were allowed food and water ad libitum. All the research work carried out was in accordance with the Institutional Review Board (IRB) of the University of Lahore.
The rats were then randomly divided into six groups with five rats (n=5) in each group. Group 1 was labeled as negative controls as they received a normal diet, while diabetes was induced in the rest of the groups through intraperitoneal injection of alloxan at 150 mg/kg body weight. Group 2 was labeled as positive control since no treatment was introduced. Group 3 and group 4 were administered iso-orientin at 200 mg/kg and lupeol at 200 mg/kg respectively through oral administration (by adding it to their food). Group 5 rats were administered the combination of lupeol and iso-orientin, whereas group 6 was administered with commercially available insulin.
Ethical statement
Ethical and legal approval was obtained from the IRB, The University of Lahore before the commencement of the study and all experiments were performed according to the guidelines of the IRB, The University of Lahore.
Collection of blood and serum separation
Following the various treatments, after a few weeks the subjects were followed for their blood glucose levels and were aymptomised as subjects before collection of their blood. An amount of 5 mL of blood was drawn from the rats after dissection. Serum was separated by centrifugation at 3,000 rpm for 10 minutes. Serum samples were then stored at −80°C for further analysis.
Biochemical assay
Blood glucose levels were estimated by using commercially available digital blood glucose analyzer and its strips. Certain stress markers were measured to evaluate oxidative stress. Malondialdehyde (MDA) was estimated in terms of thiobarbituric acid reactive substances with the help of spectrophotometric method as described by Ohkawa et al.14 Superoxide dismutase (SOD) was estimated with the help of another spectrophotometric method described by Kakkar et al.15 Glutathione (GSH), glutathione peroxidase (GPx), glutathione reductase (GRx), catalase (CAT), and nitric oxide (NO) were estimated by their respective spectrophotometric methods as explained by Moron et al,16 Sweazea et al,17 and Bredt and Synder.18
Structure prediction
The amino acid sequences of selected rat proteins were retrieved in FASTA format from Uniprot Knowledgebase database19 and were subjected to BLASTp for the identification of suitable templates against PDB. The utilized parameters were set according to the nature of the query. Alignment was done by a compositional matrix adjust scoring system. The alignment was done as the requirement of model building through MODELLER 9.15.20 The templates were retrieved considering the e-value, identity, and query coverage for structure modeling. Comparative modeling and threading approaches were employed to predict 3D structures by using MODELLER 9.15, I-Tasser,21 Modweb,22 EsyPred,23 3D-JigSaw,24 and SwissModel25 which are based on spatial restraints. Numerous evaluation tools [ANOLEA (Atomic Non–Local Environment Assessment),26 Rampage,27 ERRAT,28 and Molprobity]29 were used to evaluate the predicted protein structures.
Model evaluation was done for all generated structures including models through MODELLER 9.15 and web-based servers. Evaluation tools were applied to determine the accuracy of predicted structures. Rampage generated a Ramachandran plot and provided information about favored, allowed, and outlier regions. ERRAT evaluated the overall quality of the predicted structures. The selected structure was subjected to energy minimization by employing UCSF Chimera 1.8 and utilized steepest descent and conjugate gradient at 1,000 runs with Amber force-field parameters.
Molecular docking
The geometry optimization and energy minimization of selected molecules were performed by Chem3D Ultra, Avogadros, and UCSF Chimera 1.10. Docking studies were conducted to determine the interactions and principles of bonding between proteins and ligands. Docking was performed by using AutoDock-Vina at 1,000 runs.30 The generated results were analyzed and visualized by UCSF Chimera 1.1131,32 and Lig-Plot.33
Identification of active sites was analyzed from the literature using various tools: CASTP,34 GALAXYsite,35 COACH,36 RaptorX binding,37 and 3D-ligandsite.38 ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties were calculated from admetSAR online server,39 while the Lipinski’s rule of five was examined by using a mCule server.40
Results
Results of the study demonstrated higher blood glucose levels in rats receiving alloxan (intraperitoneally) (208±6.94 mg/dL) as compared to normal rats (90±7.38 mg/dL), as shown in Table 1, indicating that an experimental diabetic condition was successfully induced. Rats provided with a single therapeutic dose of lupeol and iso-orientin showed a reduction in their blood glucose levels (138.26±10.26 mg/dL and 156.15±7.26 mg/dL) respectively. Combined treatment of both extracts, ie, lupeol and iso-orientin led to significant synergistic results (129.06±6.29 mg/dL). Likewise, rats administered insulin had an even greater reduction in their blood glucose levels, which were very close to the healthy rats.
 | Table 1 Antioxidative antioxidant profile of rats receiving alloxan vs controls |
Similarly, levels of the different oxidants and antioxidants showed significant changes in the group with alloxan-induced diabetes. Levels of MDA in alloxan group were higher (4.06±1.09 µmol/L) as compared to controls (1.06±0.005 µmol/L), whereas, treatment with lupeol and iso-orientin significantly reduced MDA levels to 2.01±0.056 µmol/L and 1.96±0.041 µmol/L, respectively. Administration of lupeol and iso-orientin in combined form reduced level of MDA to 1.61±0.013 µmol/L. As shown in Table 1, the levels of SOD, GSH, and CAT were significantly reduced in rats which received alloxan as compared to healthy ones. Significant changes were observed after the administration of these extracts, ie, individual and combined administration of lupeol and iso-orientin significantly ameliorated the levels of discussed antioxidants. Enzymes of GSH such as GPx and GR play a key role in conversion of glutathione into its oxidized or reduced forms. Diabetes-induced rats had significant reduction in levels of GPx and GR. Combination of both lupeol and iso-orientin showed positive effects in reducing these levels.
Alloxan decreased rats' body weight significantly (168.18±2.55 g) as compared to healthy controls (245.16±5.26 g), this effect may have been ameliorated by introduction of both lupeol and iso-orientin, as shown in Table 1, as body weight of rats receiving these extracts (205.56±3.55 g) remained higher than untreated diabetic rats (168.18±2.55 g). Similarly, alloxan increased the production of reactive nitrogen species (RNS) by the up-regulation of levels of NO that were reported as 33.26±5.23 µmol/L in rats receiving alloxan compared to healthy rats (18.26±4.26 µmol/L). Synergistic effects of discussed extracts can be observed in the case of NO – they significantly decreased the levels of NO (22.25±3.25 µmol/L) as compared to rats receiving alloxan (33.26±5.23 µmol/L). Therefore, these extracts of lupeol and iso-orientin were subjected to 3D structure generation, as shown in Table 2. Six prepared energy minimized proteins were subjected to these two ligands, as shown in Figure 1, and docked structures are discussed in the following section.
 | Table 2 Templates of targeted proteins stored by their overall quality, query coverage, E-values, and identity |
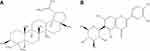 | Figure 1 Schematic representation of the two-dimensional chemical structures (A) lupeol and (B) iso-orientin. |
Structure prediction
The sequences of selected proteins (aldose reductase, glucose-6-phosphatase, glycogen phosphorylase, GPR40, GLUT-4, UCP2) were subjected to BlastP against PDB database to search for suitable templates against the target sequences. The top-ranked, optimally aligned, suitable templates for each protein with query coverage, maximum identity, E-values, and total scores were selected for homology modeling. The scrutinized templates were utilized to generate 3D structures of selected proteins. The overall query coverage and similarity among the utilized templates and target showed >65% from end-to-end, which was not considered satisfactory for reliable structures using homology modeling approach for all the selected proteins except for glucose-6-phosphatase.
Threading approach was utilized for the 3D structure prediction of glucose-6-phosphatase. Fifteen models for each selected protein were predicted by utilizing numerous tools (3D-jigsaw, Swiss Model, I-TASSER, and MODELLER 9.13) based on in silico approaches (homology modeling and threading) by satisfying the sequence and structural information from literature. All the generated models were evaluated on the basis of favored region, allowed region, outliers, overall quality factor (Figure S1), and binding regions. The most reliable structure was selected for further studies. The selected structures, after the critical examination of evaluation parameters, were subjected to UCSF Chimera 1.8 for minimization at 1,000 steepest and conjugate gradient runs. The selected minimized structure of selected proteins (Figure 2) has potential to be employed in further molecular docking studies.
Molecular docking analyses
Based on extensive literature studies, two phytochemicals (lupeol and iso-orientin) were investigated for identification of binding domains by conducting molecular docking analysis of the selected proteins. Nevertheless, the comparative docking analyses of scrutinized ligands revealed variations in their binding energies. Initially, the molecular docking analyses were performed with 100 runs and all the generated docking complexes were analyzed, out of which the suitable docked complex with the lowest binding energy and highest binding affinity was chosen for each protein. Docking analyses were conducted against the selected phytochemicals by utilizing AutoDock Vina docking software. The utilized docking tools showed effective binding energies and also similar binding residues.
The ligands were critically analyzed on the basis of their lowest binding energy values, highest binding affinity, and drug properties. The lowest binding affinities were calculated (−8.9 to −12.6 Kcal/mol) and interactive residues were also enlisted (Table 3). The lowest binding energy was observed in GLUT-4. Interestingly, it was also observed that the utilized ligands showed effective binding where the docked complex was observed, as shown in Figure 3. The Lipinski’s rule of five was calculated by mCule server (Table 4). Iso-orientin showed the lowest binding affinity (−12.6 Kcal/mol) against GLUT-4 and lupeol showed the lowest binding affinity (−10.4 Kcal/mol) against UCP-2.
 | Table 3 Calculation of binding affinities and interactive residues in the docked complex |
 | Table 4 mCule properties of ligands according to Lipinski's rule of five |
Discussion
The present study is of great significance as it is the first one to report in silico findings of lupeol and iso-orientin. It indicates their potential role as antidiabetic agents. Worldwide, diabetes mellitus is recognized as the third most common and serious metabolic disease that can even cause death. Apart from the efficacy of insulin and several oral hypoglycemic drugs used for the treatment of diabetes mellitus, they still have a number of side effects. Management of this metabolic disorder has a number of challenges for both patients and physicians.41 Investigations have revealed that traditional herbal medicines offer exciting opportunities, such as unlimited resources for the discovery of new drugs that can be used for the welfare of mankind. Moreover, herbal medicines can be used to treat a number of metabolic disorders like tumor invagination, oral disorders, and diabetes mellitus due to hypoglycemic activities etc. With all such approaches herbal medicines have relatively fewer side effects than modern medicine.42
Superoxide anion radicals (O2), hydrogen peroxide (H2O2), and hydroxyl radicals (OH) may be considered as major ROS that are generated during the onset of increased oxidative stress. These species play a significant role in the damage of pancreatic beta cells. Normally, aerobic cells are endowed with extensive mechanisms that include low molecular weight scavengers such as decreased GSH, ascorbic acid (vitamin C), vitamin E, and enzymes namely SOD, CAT, and GPx. These antioxidants play an essential role in eliminating the ROS produced during normal physiological functions.
It also helps to explain the lethal effects of alloxan in inducing diabetes and oxidative stress. Alloxan can increase levels of MDA and NO in a rat model. Similarly, CAT catalyses the decomposition of H2O2 into H2O and O2. In diabetes, the activity of CAT is decreased in the low availability of NADPH or gradual reduction in the erythrocytes, where alloxan decreases the levels of CAT and other antioxidants that cause oxidative stress. GPx reduces H2O2 to H2O. Following lupeol and iso-orientin treatments, the activity of SOD was ameliorated in diabetic rats, as also seen for CAT, GSH, and GPx. Increased levels of lipid peroxidation in rats resulted in elevation of MDA in the biological system. A higher level of NO results in an increased level of peroxynitrite in the body, which leads to increased RNS levels. Alloxan-induced diabetes also caused oxidative stress by reducing the levels of antioxidants such as GSH, SOD, CAT, GPx, and GR in the serum of diabetic rats.
In the current study, plant-derived phytocompounds (lupeol and iso-orientin) were used against six target proteins (GPR40, glucose-6-phosphatase, UCP2, glycogen phosphorylase, aldose reductase, and GLUT-4) to detect their efficacy against diabetic disorders. For aldose reductase, the key residues were identified (Try-20, Tyr-48, His-110, Trp-111, Phe-122, Tyr-219, Leu-300, Ser-302 and Cys-303) from literature.43 The previous study indicated that Tol ligand binds with aldose reductase which also interacts with these residues. In this study, that particular structure was mutated as compared to reported residues. The concerned ligands were bound with aldose reductase in a similar position as Tol ligand.
Current docking, as shown in Figure 4, of aldose reductase revealed that the particular residues are involved in mutant structure with Val-48 in place of Tyr-48, His-111 in place of Trp-111, Pro-219 in place of Trp-219, and Met-302 in place of Ser-302. The active sites for glucose-6-phosphatase, all that key binding sites (Arg-79, Arg-83, His-114, His-119, His-167 and His-176) interacted with glucose-6-phosphatase-β.44 In the current study, the residues which strongly interacted with glucose-6-phosphatase were identified (Trp-63, Val-67, Trp-70, Thr-194, Phe-195, Phe-219, Phe-223, Leu-226, Leu-273, Asn-276, Tyr-280, Pro334, Val-338). The key residues Gly-135, Leu-136, Leu-139, Asp-283, Asn-284, Asp-339, His-377, Thr-378, Val-455, Asn-484, Tyr-573, Lys-574, Glu-672, Gly-675, and Thr-676 were identified as active sites for glycogen phosphorylase. In the current study, the ligands did not bind at similar positions as in previously reported protein structures.45 The CP4, PLP, NBG, and MPD ligands also interacted at different positions with glycogen phosphorylase.46 The current in silico study revealed that the following key interactive residues interacted with glycogen phosphorylase: Met-1, Gln-72, Arg-94, Gln-97, Gln-106, Asp-110, Tyr-114, Asp-119, Met-120, Glu-121, Glu-124, Glu-125, Tyr-156, Arg-194, Phe-197, Arg-310, Arg-311, Leu-495, Cys-496, Pro-498, Lys-545 and Lys-656.
 | Figure 4 The molecular interactions of top-ranked docked complexes as analyzed by Lig-Plot. |
As for the molecular docking analyses of GPR40, in a previous study Srivastava et al, in 2014, established that Pro-80, Val-81, Ala-83, Val-84, Phe-87, Tyr-91, Leu-135, Leu-1368, Gly-139, Gly-143, Phe-142, Val-141, Leu-171, Glu-172, Trp-174, Ala-179, Ala-182, and Arg-183 had molecular interactions with GPR40 at allosteric sites. In this study, all ligands bound to GPR40 at the allosteric site positions similar to those reported in the literature.47
Thr-30, Gln-161, Ile-164, Val-165, Ile-168, Gln-282, Gln-283, Ile287, Asn-288, Phe-291, Asn-317, Pro-380, Gly-384, Pro-385, Try-388, Phe389, Asn-411, and Asn-415 were observed as key binding sites for GLUT-4 based on literature.48 The B-nonylglucoside ligand bound with GLUT-1 and in the current study both ligands bound at a similar position as that of the reported ligand. In UCP2, the active site was not reported in literature.49 The active sites were identified by utilizing different tools, ie, CASTP, GALAXYsite, COACH, RaptorX binding, and 3D-ligandsite. Our current docking study on UCP-2 indicated strong interaction between both ligands where some key residues (Ala-12, Ala-15, Asp-16, Thr-19, Phe-20, Asp-23, Lys-26, Arg-76, Val-130, Gln-133, Asn-170, Asp-224, Lys-227, and Tyr-230) were involved in strong interactions. To the best of our knowledge, our study is the first to report on UCP-2 docking where both ligands were found to bind at similar positions and all molecular interactions strongly interacted with the targeted UCP-2 protein.
The results of docking analyses were considered satisfactory for drug designing and both ligands presented effective results for targeting diabetes. Docking results of lupeol and iso-orientin showed strong activity with all six target proteins and the difference between binding energies can be observed in Table 5. Both ligands shared common residues for potent inhibition of protein activity in diabetes. In this study, lupeol and iso-orientin were shown to possess significant antidiabetic activity based on the ADMET profile.
 | Table 5 Calculation of ADMET profile |
The blood–brain barrier (BBB) is highly significant in terms of drug transport across the brain and is a very efficient barrier that is designed to filter molecules which may be harmful to brain function.50 Intestinal absorption is an important factor for effective drug candidates and human intestinal absorption properties depend on the physiochemical properties of compounds.51 Better ADMET profile analysis was seen in the case of lupeol as compared to iso-orientin. Lupeol is also more potent as it has the ability to cross the BBB.
One of the essential properties of phytochemicals is biodegradation. It is a factor of dissipation into the environment and is also known as biotransformation. Phytochemicals which are not readily or efficiently degraded may be responsible for the toxicity in terrestrial and aquatic ecosystems due to the production of CO2 and reactive intermediates.39 The biodegradation principle is determined by various experimental measurements including the production of CO2, biochemical oxygen demand, pharmacokinetic properties, the development of stable constants, and the formation of reactive intermediates.52 In silico approach is a principle for assessing biodegradability, bioavailability, BBB, drug toxicity and carcinogenicity of compounds. Our study indicated that both phytochemicals lack the biodegradation property. In silico ADMET profile analysis is an efficient and rapid procedure to determine the pharmacokinetic properties of compounds. Better ADMET profile analysis was seen in the case of lupeol, which is also more potent than iso-orientin due to its ability to cross the BBB. In addition, although both compounds showed low CYP inhibitory promiscuity, they were resistant to biodegradation, were non-AMES toxic, and non-carcinogenic. Therefore, lupeol and iso-orientin have the potential to be investigated as antidiabetic-agents.
Due to the following limitations: financial constraints, socio-economic obstacles, limited resources etc, the current study was conducted with a small sample size and limited variables were used. Therefore, in future, a study may be performed with a larger sample size, with assessment of other important variables (AGEs, AOPPs, and MMPs) which could lead to evaluation of the mechanism of action of antidiabetic effect of plant extracts before proposing it as a new antidiabetic medicine.
Conclusion
The current study revealed the antidiabetic activity of discussed phytocompounds (lupeol and iso-orientin). It showed that administration of these compounds, either individually or in combined form, significantly reduced the levels of blood glucose and ameliorated the antioxidative profile of the body. Therefore, further clinical studies on human models, using these compounds, may help to elucidate their significant therapeutic benefits in diabetes and some other disorders.
Acknowledgments
The authors are very thankful to students and staff of Lab-313, Institute of Molecular Biology and Biotechnology, The University of Lahore, Pakistan for providing innovation and support for the project.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rahmani AH. Cassia fistula linn: potential candidate in the health management. Pharmacognosy Res. 2015;7(3):217. doi:10.4103/0974-8490.157956
2. Srividhya M, Hridya H, Shanthi V, Ramanathan K. Bioactive amento flavone isolated from Cassia fistula L. leaves exhibits therapeutic efficacy. 3 Biotech. 2017;7(1):33. doi:10.1007/s13205-017-0599-7
3. Sangu PK, Krishna MC, Sharma BK, Rajashekharan R, Prasad GP, Ala N. Medicinal properties of Aragvadha (cassia fistula Linn.). Int J Ayurvedic Med. 2010;1(3):129–133.
4. As K. Leguminous trees and their medicinal properties. In: Medicinally Important Trees. Cham: Springer; 2017:211–233.
5. Patel DK, Kumar R, Laloo D, Hemalatha S. Diabetes mellitus: an overview on its pharmacological aspects and reported medicinal plants having antidiabetic activity. Asian Pac J Trop Biomed. 2012;2(5):411–420. doi:10.1016/S2221-1691(12)60067-7
6. Asmat U, Abad K, Ismail K. Diabetes mellitus and oxidative stress—a concise review. Saudi Pharm J. 2016;24(5):547–553. doi:10.1016/j.jsps.2015.03.013
7. Fan W. Epidemiology in diabetes mellitus and cardiovascular disease. Cardiovasc Endocrinol Metab. 2017;6(1):8–16. doi:10.1097/XCE.0000000000000116
8. Rufinatscha K, Radlinger B, Dobner J, et al. Dipeptidyl peptidase-4 impairs insulin signaling and promotes lipid accumulation in hepatocytes. Biochem Biophys Res Commun. 2017;485(2):366–371. doi:10.1016/j.bbrc.2017.02.071
9. Rameshreddy P, Uddandrao VS, Brahmanaidu P, et al. Obesity-alleviating potential of asiatic acid and its effects on ACC1, UCP2, and CPT1 mRNA expression in high fat diet-induced obese Sprague–dawley rats. Mol Cell Biochem. 2018;442(1–2):143–154. doi:10.1007/s11010-017-3199-2
10. Qasim M, Abideen Z, Adnan MY, et al. Antioxidant properties, phenolic composition, bioactive compounds and nutritive value of medicinal halophytes commonly used as herbal teas. S Afr J Bot. 2017;110:240–250. doi:10.1016/j.sajb.2016.10.005
11. Salem N, Bachrouch O, Sriti J, et al. Fumigant and repellent potentials of ricinus communis and mentha pulegium essential oils against tribolium castaneum and lasioderma serricorne. Int J Food Prop. 2018;11:1–5.
12. Sehgal SA, Khattak NA, Mir A. Structural, phylogenetic and docking studies of D-amino acid oxidase activator (DAOA), a candidate schizophrenia gene. Theor Biol Med Model. 2013;10(1):3. doi:10.1186/1742-4682-10-3
13. Bhalerao SA, Kelkar TS. Traditional medicinal uses, phytochemical profile and pharmacological activities of cassia fistula linn. Int Res J Biol Sci. 2012;1(5):79–84.
14. Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95(2):351–358.
15. Kakkar P, Das B, Viswanathan PN. A modified spectrophotometric assay of superoxide dismutase (SOD). Indian J Biochem Biophys. 1984;21:131–132.
16. Moron MS, Depierre JW, Mannervik B. Levels of glutathione, glutathione reductase and glutathione S-transferase activities in rat lung and liver. Biochimica et Biophysica Acta (BBA)-Genl Sub. 1979;582(1):67–78. doi:10.1016/0304-4165(79)90289-7
17. Sweazea KL, Johnston CS, Knurick J, Bliss CD. Plant-based nutraceutical increases plasma catalase activity in healthy participants: A small double-blind, randomized, placebo-controlled, proof of concept trial. J Diet Suppl. 2017;14(2):200–213. doi:10.1080/19390211.2016.1207742
18. Bredt DS, Snyder SH. Transient nitric oxide synthase neurons in embryonic cerebral cortical plate, sensory ganglia, and olfactory epithelium. Neuron. 1994;13(2):301–313.
19.
20. Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr protoc protein sci. 2016;86(1):2–9. doi:10.1002/cpps.20
21. Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7. doi:10.1038/nmeth.3213
22. Pieper U, Webb BM, Dong GQ, et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2013;42(D1):D336–46. doi:10.1093/nar/gkt1144
23. Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics. 2002;18(9):1250–1256.
24. Bates PA, Kelley LA, MacCallum RM, Sternberg MJ. Enhancement of protein modeling by human intervention in applying the automatic programs 3D‐JIGSAW and 3D‐PSSM. Proteins. 2001;45(S5):39–46. doi:10.1002/prot.1168
25. Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(W1):W252–8. doi:10.1093/nar/gku340
26. Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. InISMB. 1997;5:187–190.
27. Lovell SC, Davis IW, Arendall WB, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins. 2003;50(3):437–450. doi:10.1002/prot.10286
28. Lengths MC, Angles MC. Limitations of structure evaluation tools errat. Quick Guideline Comput Drug Des. 2018;16:75.
29. Chen VB, Arendall WB, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Sect D. 2010;66(1):12–21. doi:10.1107/S0907444909042073
30. Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
31. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi:10.1002/jcc.20084
32. Kosinski J, von Appen A, Ori A, Karius K, Müller CW, Beck M. Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J Struct Biol. 2015;189(3):177–183. doi:10.1016/j.jsb.2015.01.014
33. Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to ser‐His‐Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996;5(6):1001–1013. doi:10.1002/pro.5560050603
34. Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(suppl_2):W116–8. doi:10.1093/nar/gkl282
35. Heo L, Shin WH, Lee MS, Seok C. GalaxySite: ligand-binding-site prediction by using molecular docking. Nucleic Acids Res. 2014;42(W1):W210–4. doi:10.1093/nar/gku321
36. Yang J, Roy A, Zhang Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29(20):2588–2595. doi:10.1093/bioinformatics/btt447
37. RaptorX Binding [homepage on the Internet]. To identify binding region of protein have used online tool. Accessed March 20, 2018.
38. Wass MN, Kelley LA, Sternberg MJ. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38(suppl_2):W469–73. doi:10.1093/nar/gkq406
39. Cheng F, Li W, Zhou Y, et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012;52:3099–3105.
40. Kiss R, Sandor M, Szalai FA. http://Mcule. com: a public web service for drug discovery. J Cheminform. 2012;4(S1):P17. doi:10.1186/1758-2946-4-S1-P17
41. Ghosh JR, Dastidar PG, Dey B, Das P, Bandyopadhyay AR. Palmar dermatoglyphic traits in type 2 diabetes mellitus patients of Bengalee Hindu caste population of West Bengal, India: a cross-sectional study. J Biomed Sci. 2018;3(2):18–23. doi:10.3126/jbs.v3i2.18919
42. Hussain F, Rana Z, Shafique H, Malik A, Hussain Z. Phytopharmacological potential of different species of Morus alba and their bioactive phytochemicals: a review. Asian Pac J Trop Biomed. 2017;7(10):950–956. doi:10.1016/j.apjtb.2017.09.015
43. Steuber H, Heine A, Podjarny A, Klebe G. Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features. J Mol Biol. 2008;379(5):991–1016. doi:10.1016/j.jmb.2008.03.063
44. Tang W, Martin KA, Hwa J. Aldose reductase, oxidative stress, and diabetic mellitus. Front Pharmacol. 2012;9(3):87.
45. Anderka O, Loenze P, Klabunde T, et al. Thermodynamic characterization of allosteric glycogen phosphorylase inhibitors. Biochemistry. 2008;47(16):4683–4691. doi:10.1021/bi702397d
46. Rath VL, Ammirati M, Danley DE, et al. Human liver glycogen phosphorylase inhibitors bind at a new allosteric site. Chem Biol. 2000;7(9):677–682. doi:10.1016/S1074-5521(00)00004-1
47. Srivastava A, Yano J, Hirozane Y, et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513(7516):124. doi:10.1038/nature13494
48. Deng D, Xu C, Sun P, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510(7503):121. doi:10.1038/nature13306
49. Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476(7358):109. doi:10.1038/nature10257
50. Chen Y, Liu L. Modern methods for delivery of drugs across the blood–brain barrier. Adv Drug Deliv Rev. 2012;64(7):640–665. doi:10.1016/j.addr.2011.11.010
51. Augustijns P, Wuyts B, Hens B, Annaert P, Butler J, Brouwers J. A review of drug solubility in human intestinal fluids: implications for the prediction of oral absorption. Eur J Pharml Sci. 2014;57:322–332. doi:10.1016/j.ejps.2013.08.027
52. Dimitrov S, Pavlov T, Nedelcheva D, et al. A kinetic model for predicting biodegradation. SAR QSAR Environ Res. 2007;18(5–6):443–457. doi:10.1080/10629360701429027
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



