Back to Journals » Drug Design, Development and Therapy » Volume 12
Identification of novel drug targets in bovine respiratory disease: an essential step in applying biotechnologic techniques to develop more effective therapeutic treatments
Authors Sakharkar MK , Rajamanickam K , Chandra R, Khan HA , Alhomida AS, Yang J
Received 23 January 2018
Accepted for publication 14 March 2018
Published 7 May 2018 Volume 2018:12 Pages 1135—1146
DOI https://doi.org/10.2147/DDDT.S163476
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tuo Deng
Meena Kishore Sakharkar,1 Karthic Rajamanickam,1 Ramesh Chandra,2 Haseeb A Khan,3 Abdullah S Alhomida,3 Jian Yang1
1College of Pharmacy and Nutrition, University of Saskatchewan, Saskatoon, SK, Canada; 2Department of Chemistry, University of Delhi, Delhi, India; 3Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
Background: Bovine Respiratory Disease (BRD) is a major problem in cattle production which causes substantial economic loss. BRD has multifactorial aetiologies, is multi-microbial, and several of the causative pathogens are unknown. Consequently, primary management practices such as metaphylactic antimicrobial injections for BRD prevention are used to reduce the incidence of BRD in feedlot cattle. However, this poses a serious threat in the form of development of antimicrobial resistance and demands an urgent need to find novel interventions that could reduce the effects of BRD drastically and also delay/prevent bacterial resistance.
Materials and methods: We have employed a subtractive genomics approach that helps delineate essential, host-specific, and druggable targets in pathogens responsible for BRD. We also proposed antimicrobials from FDA green and orange book that could be repositioned for BRD.
Results: We have identified 107 putative targets that are essential, selective and druggable. We have also confirmed the susceptibility of two BRD pathogens to one of the proposed antimicrobials – oxytetracycline.
Conclusion: This approach allows for repositioning drugs known for other infections to BRD, predicting novel druggable targets for BRD infection, and providing a new direction in developing more effective therapeutic treatments for BRD.
Keywords: BRD, pathogenic bacteria, targets, drugs, prioritization, differential genome analyses, druggability
Introduction
The most prevalent infectious disease experienced by stockers, producers, and feedlot cattle is bovine respiratory disease (BRD).1 BRD has deleterious effects on cattle health and performance resulting in considerable economic loss.2–6 BRD is caused by multiple factors, including a combination of bacterial and viral components.7,8 Associated factors such as environmental and stress-related exposures (eg, weaning and transportation) also play a part.9–14 According to data provided by the Canadian Cattlemen Association, BRD accounts for 65%–80% of the sickness in some feedlots, 45%–75% of the death loss, and an annual loss of about US$ 600–750 million to the North America beef industry.15 The key pathogenic bacteria for BRD are Mannheimia hemolytica, Haemophilus somni, and Pasturella multocida in Canada.16,17 Although not of major concern, these pathogens are also reported to be involved in other infections in cattle, such as mastitis.18,19 Vaccination has shown inconsistent results in terms of protection against BRD pathogens.20 Consequently, primary management practices such as metaphylactic antimicrobial injections for BRD prevention are used to reduce the incidences of BRD in feedlot cattle.21 These practices may contribute to antimicrobial resistance (AMR), which, in turn, will reduce the efficacy of the antimicrobials commonly employed to control infectious disease in cattle.22 To this end, there is a need to develop an effective treatment protocol for BRD that is efficacious, reduces antimicrobial usage, and effectively manages the development of resistance.23
An essential step in developing any novel therapeutic treatment is target identification and early validation.22 With the large amount of data on pathogenic bacterial genomes, genomics can be applied to evaluate the suitability of potential targets using two criteria, “essentiality” and “selectivity”.24–26
The target must be essential for the growth, replication, viability, or survival of the microorganism, that is, encoded by genes critical for pathogenic life-stages.27 Essential genes that constitute the foundation of life of the microorganism are critical for survival, and, therefore, are likely to be common and conserved in all bacterial species.28–31 Disruption of these genes can cause lethality, making them attractive drug targets.
The microbial target for treatment should not have any well-conserved homolog in the host, that is, it should be “selective” in order to minimize cytotoxicity issues.24,28 Thus, only genes present in the pathogenic genome and absent from host genome can be considered as candidate drug targets. For selectivity, subtractive genome analysis has been employed as an important technique and has contributed toward target identification and selection in several pathogens.32–36 This would help to avoid expensive dead-ends when a lead target is identified and investigated in great detail only to discover at a later stage that all its identified inhibitors have off-target effects in the host. This approach paired with the ability to predict essential genes can help to identify potential targets for drug development.24,28
The druggability of targets is another prioritization filter that determines the potential of a prioritized target to be modulated by a small-molecule drug.37 This is important because the complete proteome data of several pathogens, supplemented by gene essentiality data and data on drugs against these pathogens and their respective mechanism of action, may help to identify essential druggable targets.
Here, we demonstrate the unprecedented potential of complementary datasets (gene essentiality, subtractive genomics, and druggability) for drug target prioritization. We employed genome matching techniques for the identification of proteins specific to the pathogen and used the Database of Essential Genes (DEG), which provides gene essentiality data for 46 bacterial pathogens to select essential genes. The DrugBank database, which is a resource that combines detailed drug data with comprehensive drug target information, was used to assign druggability to candidate targets.38 The above databases, along with completely sequenced genome data for Bos taurus and the BRD pathogens, provide a basis for using selectivity, essentiality, and druggability as criteria for addressing the complexities and conundra in target prioritization by computational methods. To better understand the cellular organization and functionality of the prioritized targets, we also analyzed the protein pathway and the gene ontology (GO) of these targets.39 Our results will be extremely beneficial in developing more effective therapeutic strategies for BRD, such as novel drug development and drug repositioning.
Materials and methods
The proteomes of the host B. taurus and the key BRD pathogens – H. somni strain 2336 (NC_010519.1), M. hemolytica strain M42548 (NC_021082.1), and P. multocida strain 36950 (NZ_CP008918.1) – were downloaded (20th October, 2017) from the National Center for Biotechnology Information (NCBI) website. The H. somni, M. hemolytica, and P. multocida proteomes have 1,957, 2,833, and 1,856 proteins, respectively. The B. taurus proteome has 49,107 proteins (Table 1).
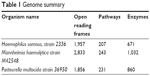  | Table 1 Genome summary |
Essential genes
As gene essentiality data were not available for the BRD pathogens, the proteomes of the BRD pathogens were subjected to Basic Local Alignment Search Tool for Proteins against the DEG protein database at an expect value (E-value) cutoff at 10−10 and bit score >100 to identify proteins that could potentially be essential and, hence, be possible drug targets. The E-value describes the number of hits one can “expect” to see by chance when searching a database.
Subtractive genome analyses
To minimize drug cross-reactivity due to binding to homologous proteins in the host, BLASTP analyses were carried out for all the three pathogens involved in BRD against the B. taurus proteome at an E-value cutoff of 10−4 and bit score >100 to exclude host proteins that are similar to the pathogen proteins. This will help to screen out proteins that are essential for BRD pathogens but have B. taurus homologs.
Druggable proteins
The druggability of screened proteins was investigated against all drug targets present in the DrugBank database, which contains 8,261 drug entries including 2021 FDA (US Food and Drug Administration)-approved small-molecule drugs, 233 FDA-approved biotech (protein/peptide) drugs, 94 nutraceuticals, and over 6,000 experimental drugs.40 Additionally, 4,338 nonredundant protein (ie, drug target/enzyme/transporter/carrier) sequences are linked to these drug entries. The resultant BLASTP hits from these two searches with a bit score >100 and an E-value cutoff of <10−5 were considered as potentially druggable therapeutic candidates. We also performed a BLASTP search of the druggable targets against 85 bovine gut microbial genome sequences at a cutoff of 10−100 and bit score >100 to identify targets that do not give a match in the gut microbiome (data not shown).
The druggable targets were subsequently subjected to quantitative analyses like pathway analyses, GO analyses, choke-point analyses, prediction of protein sorting signals and localization sites (PSORT) biologic location analyses, virulence analyses, and AMR analyses. A list of drugs approved for BRD was downloaded from the FDA Green Book to act as a positive control.
The drugs that bind to the predicted druggable targets identified above (dataset 1) were also matched with the FDA Orange Book and FDA Green Book to identify drugs that are approved on the basis of safety and effectiveness under the Federal Food, Drug, and Cosmetic Act (the Act) and related patent and exclusivity information (Table 2).
Pathway analyses
The putative druggable targets were analyzed by KAAS (kyoto encyclopedia of genes and genomes [KEGG] Automated Annotation Server) to obtain data on biologic processes and metabolic pathways.41 KAAS performs a BLASTP comparison against the KEGG genes database and provides functional annotations for target proteins.
Choke-point analyses
Choke-point analysis of the metabolic pathways of the BRD-causing organisms was conducted using the BioCyc database. A list of choke-point reactions and the respective proteins catalyzing the reactions was downloaded.41 The list of druggable putative targets was screened against this choke-point list to identify targets for which no alternate pathways were available.42
Gene ontology
GO – biologic and molecular functions and cellular distribution were assigned for the prioritized targets using the Uniprot web server (http://www.uniprot.org/).43
Subcellular localization
The PSORT beta version (PSORTb) server was used to predict the subcellular localization of the putative druggable targets to analyze the localization of these targets in the different compartments of the cell.44
Virulent factor analyses
The Virulence Factor DataBase (VFDB) (http://www.mgc.ac.cn/VFs/) is a public resource that provides current knowledge about virulence factors (VFs) from several bacterial pathogens.45 BLASTP against VFDB was executed at a cutoff of 10−5 and bit score >100 to identify putative VF from selected BRD-causing organisms.
Determination of antimicrobial susceptibility
Oxytetracycline (Alpha Acer, Canada) is a drug against one of our putative druggable targets and is FDA approved. Hence, we chose it as a drug to validate our identified targets in vitro. The experiment was carried out in duplicate. The bacterial strains of M. haemolytica ATCC 29702 and P. multocida ATCC 43137 were purchased from CEDARLANE Corporation (Burlington, ON, Canada) and revived according to the manufacturer’s instructions. The antimicrobial susceptibility to oxytetracycline was determined using a 96-well plate. The concentrations of oxytetracycline used were 8, 4, 2, 1, and 0.5 μg/mL. Pure subcultures were inoculated in a Brain Heart Infusion Broth and incubated overnight at 35°C. The bacterial suspensions were then adjusted to 0.5 McFarland turbidity standards as per the manufacturer’s instructions. The bacterial suspensions (100 μL) were then added to each well. The plate was incubated at 35°C for 24 h and was subsequently read using a 96-well plate reader (BIORAD iMark Microplate Reader 655 nm; Table 3).
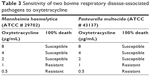  | Table 3 Sensitivity of two bovine respiratory disease-associated pathogens to oxytetracycline |
A flow chart for the process and essential genes and druggable targets shared across the three pathogens is presented in Figure 1 and Figure 2, respectively.
  | Figure 2 Venn diagram showing the shared essential genes and putative druggable targets across the three genomes. |
Results
Herein, we present the results of a novel approach for delineating target identification in the drug discovery process for BRD.
Database of essential genes
The proteomes of H. somni, M. hemolytica, and P. multocida were subjected to BLASTP against DEG to identify essential proteins, through which 1,089, 1,246, and 1,255 essential proteins were identified, respectively. Because of their vital roles in various pathways for pathogen survival, the probability of conservation of these genes among various populations and species is high.46,47
Subtractive genomics, druggability prediction, and FDA match
An efficient and fast method to identify targets that are selective to the pathogenic species and absent in the host genome is in silico subtractive genomic analysis. BLASTP analysis of the essential proteins identified above against the B. taurus proteome identified 821, 951, and 964 proteins in the genomes of H. somni, M. hemolytica, and P. multocida that have no significant match with any of the host proteins. Out of these proteins, 62, 71, and 39 were found to be hypothetical or unknown in the respective genome, and, therefore, were not considered for any further analysis. BLASTP of the remaining essential 759, 880, and 925 proteins against the Drugbank database identified 204, 230, and 240 proteins to be druggable for H. somni, M. hemolytica, and P. multocida, respectively, based on sequence similarity. Moreover, 107 proteins were identified to be conserved across all three BRD-causing bacteria, and were subject to further analyses (Figure 2). These 107 proteins are targeted by 315 drugs. Surprisingly, none of the currently approved drugs for BRD were present in this list. This is because the BRD drugs are not present in the DrugBank compendium. We further matched these 315 drugs to the FDA Orange and Green Books (https://www.fda.gov/; accessed on 20th December, 2017) to identify drugs that are approved on the basis of safety and effectiveness by FDA under the Federal Food, Drug, and Cosmetic Act and related patent and exclusivity information. Thirty-two of our drugs were found to be present in the FDA Orange Book and 10 in the FDA Green Book (Table 2).
Pathway analyses, GO, choke point, and VF
KAAS: the 107 common prioritized drug targets were analyzed using KAAS for pathway analyses, which revealed 62 pathway annotations.29 The distribution of these 107 proteins into different pathways is presented in Figure 3. A substantial proportion of proteins were components of ribosomes (15), involved in the biosynthesis of amino acids (15), or involved in pyrimidine metabolism (11). GO: we analyzed the GO terms for the 107 prioritized target proteins (Figure 4). The analysis revealed that translation (17), regulation of cell shape (11), cell wall organization (11), and peptidoglycan biosynthesis (11) were the most common biologic processes. A total of 129 classifications were identified, suggesting that several proteins are involved in more than one biologic process. A total of 149 classifications were identified under the molecular function and the three most abundant were ATP binding (20), metal ion binding (19), and structural constituents of ribosome (16). The proteins were categorized under 26 locations in the bacterial cell, but were mostly distributed in the cytoplasm. Choke-points: we performed a choke-point analysis on the 107 potential target proteins to identify choke-point proteins. Essentiality is almost perfectly predicted by the lack of an alternative pathway.48 Out of the 107 common drug targets that were prioritized, 26 were identified as choke-point proteins. The choke-point proteins are druggable and effective targets because the inhibition of these choke-point proteins is expected to produce a blockade in the pathway, which may create an unsustainable condition inside the bacterial cell. Hence, these proteins are predicted to be attractive proteins in their respective pathways for the design of potent inhibitors. VFs: ten proteins were identified as essential, nonhost homologs, druggable, and involved in virulence. The identified VFs were encoded by the following genes: pdxA, rfaD, relA, glmM, gmhA, coaD, lpxC, ispD, kdsA, and kdsB.43 Several virulence genes are important for bacteria to establish infection and are indirectly involved in pathogenesis.49 It has recently been argued that antivirulence drugs may generate a weaker selection for resistance as compared to other antibacterial drugs, as they neutralize the pathogen’s potential to cause infection rather than being bacteriostatic or bactericidal.50 Nonetheless, resistance to drugs against virulent factors has been reported.
Subcellular locations – PSORTb
The determination of subcellular localization of proteins, especially in the case of pathogenic species, is useful in revealing their involvement in pathogenesis.51 Proteins that are easily amenable to any form of external intervention such as cell wall and cell membrane proteins are considered to be more attractive drug targets than the cytoplasmic proteins. The distribution of the predicted subcellular localization for the 107 putative druggable targets based on PSORTb is depicted in Figure 5.41 A total of 100 out of 107 (~93%) of the proteins were predicted as cytoplasmic proteins and two of these proteins were found to be cytoplasmic membrane proteins. Cytoplasmic proteins are involved in many important metabolic processes and, hence, are very important for the physiology of the bacteria. Also, most cellular activities occur in the cytoplasm. Peng and Gao also reported that essential proteins are enriched in the cytoplasm.52 Drug delivery strategies that include the use of nanoparticles, cell-penetrating peptides, pH-responsive carriers, and endosome-disrupting agents may help to overcome the barrier created by the inner membrane in these gram-negative bacteria that prevent drugs from gaining access to cytoplasmic targets. The subcellular location of five proteins could not be assigned by PSORTb. Here it is important to mention that PSORTb is a support vector machine-based algorithm and its prediction accuracy depends on the training set. At times, it may predict and assign multiple locations to a protein or may not be able to predict or assign the protein to any location in a cell.
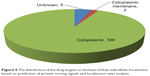  | Figure 5 The distribution of the drug targets on the basis of their subcellular localization based on prediction of protein sorting signals and localization sites analysis. |
Drugs and targets
Most of the drugs in our analysis target one protein. The drug DB08185 targets 10 proteins, which are all components of the small ribosomal subunit. Both gene gyrA and gene parC are targeted by 18 drugs, out of which 17 drugs are shared. These drugs are DB00218 – moxifloxacin (fluoroquinolone – FDA); DB00365 – grepafloxacin (quinolone – withdrawn), DB00467 – enoxacin (6-fluoronaphthyridinone – FDA), DB00487 – pefloxacin (fluoroquinolone – FDA), DB00685 – trovafloxacin (fluoroquinolones – withdrawn), DB00537 – ciprofloxacin (carboxyfluoroquinoline – FDA), DB01044 – gatifloxacin (fluoroquinolone – FDA), DB00978 – lomefloxacin (fluoroquinolone – FDA), DB01059 – norfloxacin (fluoroquinolone – FDA), DB01137 – levofloxacin (fluoroquinolone – FDA), DB01155 – gemifloxacin (fluoroquinolone – FDA), DB01165 – ofloxacin (fluoroquinolone – FDA), DB01208 – sparfloxacin (fluoroquinolone – FDA), DB01405 – temafloxacin (fluoroquinolone – withdrawn), DB04576 – fleroxacin (fluoroquinolone – FDA), DB06771 – besifloxacin (fluoroquinolone – FDA), DB09047 – finafloxacin (fluoroquinolone – FDA). DB00827 – cinoxacin (synthetic antimicrobial related to oxolinic acid and nalidixic acid – withdrawn) – only targets gyrA but not parC and DB00817 – rosoxacin (quinolone derivative antimicrobial – FDA) only targets parC but not gyrA.
Antimicrobial susceptibility
It was observed that both M. hemolytica and P. multocida are susceptible to the antimicrobial oxytetracycline (Table 3).
Discussion
BRD is a significant threat associated with morbidity and mortality in cattle and is an important problem in cattle production. In North America, an estimated USD $54.12 million per year is spent on treating cattle for respiratory disease. This does not include production losses because of morbidity and mortality.1,3,4,6 Exposure to various physical and physiologic stressors and certain viral infections predisposes the cattle to BRD.53 Several factors can contribute to BRD, which include, but are not limited to, the feedlot environmental, social, and relocation challenges and the complex interactions among the host immune system and pathogens.53–55 As the effective treatment of a disease depends on an accurate diagnosis and understating of factors that lead to it, there is poor efficiency of vaccination and antimicrobial treatments against BRD-associated bacteria.2,9,56,57
Antimicrobial metaphylaxis is the mass medication of a group of animals to eliminate or minimize an expected outbreak. Metaphylaxis is shown to reduce morbidity and mortality in feedlot and stocker arrival to cattle considered at high risk for the development of clinical BRD signs.55,58–63 However, the inability to subjectively identify, pull, and treat sick calves arriving at a feedlot or stocker facility poses a serious threat in the form of the development of AMR and warrants an urgent need to find novel interventions that could possibly reduce the effects of BRD dramatically and also delay/prevent bacterial resistance. Metaphylaxis has been reported to cause multiple drug resistance but this has not been investigated in detail.58,64
Here, it is noteworthy to mention that as BRD commonly involves a combination of pathogens and determining the specific pathogen cohort present in a newly arrived feedlot cattle is not practical, the newly identified/developed drug candidates should be against a target that can be seen across a wide range of bacteria. Essential genes have a lethality phenotype and the proteins encoded by them are important for growth and survival of the organism. They are also highly conserved across organisms and, hence, present a viable target for drugs attempting to target a wide spectrum of bacteria.29–31,65 It is important to mention that even though it is known that the selectivity of the target and the absence of the target in the host genome decrease the chances of side effects, these criteria are rarely achieved with confidence in a real sense.25 This is because the rumen microbiome has a plethora of beneficial bacteria and archaea and it is likely for a drug to target the proteins encoded by essential genes in the gut microbiome. In line with this, a BLASTP match of our prioritized 107 targets against 85 bovine gut bacterial and archeal genomes (identified from NCBI and Hunmicrobiome) showed that all the 107 proteins had significant matches with a cutoff value of 10−100 or less (data not shown). This reemphasizes the fact that antimicrobials need to be administered with caution and only when required for treating infections in cattle as they may affect/alter rumen gut flora.66
Druggability – the likelihood of being able to modulate a target by a drug is crucial in determining whether a drug discovery project progresses from a “hit” to a “lead” as druggability information guides drug discovery efforts on proteins that offer better prospects as targets.67 In view of this, druggability predictions are important to avoid expensive dead-ends and intractable targets. In the current study, we have adopted a genomics approach for drug target prioritization for delineating novel essential druggable targets for three key BRD pathogenic bacteria, H. somni, M. hemolytica, and P. multocida. A final list of 107 essential and druggable targets was obtained. These targets are involved in essential biologic processes (Figures 2 and 3). Based on the Drugbank database, 315 drugs are available for these targets. Several of the drugs target multiple pathways. However, 248 out of 315 drugs are single-target drugs, and 40 out of 107 targets are targeted by a single drug. The proteins encoded by genes gyrA and parC are targeted by the maximum number of drugs and also share the maximum number of drugs that target both of them. Most of the drugs are quinolones and their derivatives, which are reported to target essential bacterial enzymes DNA gyrase (gyrA) and topoisomerase IV (parC) as these enzymes have a high sequence identity and structural homology. Unfortunately, none of the drugs currently used for treating BRD infections in cattle (Table 2) were present in these 315 drugs. A thorough check in the DrugBank database shows that although it has several drugs used for veterinary purposes, the drugs used for BRD are missing from the database. However, it must be highlighted that a compilation of BRD drugs and their mechanism of action from the FDA Green Book and other literature shows that the key targets are DNA gyrase, the 50S ribosomal subunit, and the inhibitors of cell wall synthesis (Table 2). Interestingly, 10 of the drugs in our list are present in FDA Green Book and are approved for veterinary purposes according to the DrugBank and can be repurposed for BRD infections in cattle. Some of the drugs in the FDA Orange Book are approved for veterinary purposes and can safely be used to treat cattle infections. It is important to mention that antimicrobials that are used in human medicine may not be good choices or even approved for treatment of veterinary infections such as BRD, as there is a move away from using drugs that are important in human medicine in food-producing animals. In line with this, it is also important to mention that toxicity issues are responsible for nearly 30% of failures in drug development programs.68 Based on our results, we propose that the 10 and three antimicrobials in the FDA Green Book and Orange Book (that are approved for veterinary use), respectively, profiled for repurposing for BRD treatment (Table 2). Also, as these drugs are in the FDA and Drugbank lists and have known pharmacokinetic and safety profiles, they have a greater chance of success, pose less risks, and are economical and less time consuming as drug development targets. Furthermore, it is important to mention that <15% of compounds that enter clinical development receive approval and drug repurposing helps us to overcome this limitation in some way.69,70 To delineate drug development (including repurposing) opportunities arising from these analyses, we connected information on multiple datasets such as choke points, virulence, and GO to these druggable conserved and essential targets.
Oxytetracycline has been approved for the treatment of pneumonia and BRD associated with Pasteurella spp.71 Our current findings concur with previous reports that oxytetracycline is also effective against M. hemolytica (Table 3).72 Experimental validation of other FDA drugs on BRD pathogens and the gene sets involved in bringing out the required results may help to overcome the important issue of AMR.
On a separate note, it is also important to mention that the information on FDA-approved drugs that target multiple pathways can help us to identify drugs that can be used in combination for treating BRD and other bacterial infections.
Limitations
Some limitations of our analysis, which potentially make our list of targets and drugs incomplete, need to be mentioned. First, the terminology across datasets is inconsistent. Second, the current drug target list may not be all inclusive, as the criteria employed for the selection of targets are multiple and strict. Third, the drug lists in drug databases may not be all inclusive.73 Finally, the essential genes required for growth on different media may be different and hence our list is disputable. Also, different methods may cause different detection results. For example, genes that only slow down the growth may inaccurately be chosen as essential during transposon mutagenesis methods. Essential genes present in more than one copy may also be misclassified.74–77 Extracellular proteins may also, at times, be essential for the survival of the pathogen in the laboratory. However, as they are not generally essential for the survival of the pathogen, they are not present in the list of targets identified based on a homology with known essential proteins. We did not include these targets in our list as extracellular proteins are reported to evolve faster and, hence, are more likely to mutate and contribute to the development of resistance. This analysis does not take into consideration of distant gene protein relationships, which may be missed, because the alignment scores are likely to have a low statistical significance for these distant relationships. In addition, genes without homologs in the DEG may be missed. This is because the DEG contains experimentally determined essential genes by genome-wide essentiality screens and this datum is only available for <100 pathogens.
A possible solution to overcome these limitations is to improve the annotation across datasets and make the various datasets integrated and uniform. Also, with time, an increase in genome-wide essentiality screens for various pathogens will help to improve predictions on gene essentiality.
Conclusion
The druggable target space and drug space are not all inclusive and are expanding. The data presented here demonstrate that the stepwise prioritization of proteins using simple biologic criteria can be an effective way of rapidly reducing the number of targets of interest to an experimentally manageable number. This process is an efficient way to enrich potential target genes and identify those that are critical for normal cell function.
Acknowledgments
This work is supported by an Agriculture Development Fund (ADF) from the Government of Saskatchewan and SaskMilk and MITACS, Canada grant to MKS and JY HAK would like to acknowledge the Deanship of Scientific Research at the King Saud University (Saudi Arabia) for funding the Research Group No. RGP-009.
Disclosure
The authors report no conflicts of interest in this work.
References
Griffin D. Economic impact associated with respiratory disease in beef cattle. Vet Clin North Am Food Anim Pract. 1997;13(3):367–377. | ||
Larson R. Effect of cattle disease on carcass traits. J Anim Sci. 2005;83(13 Suppl):E37–E43. | ||
Smith RA. Impact of disease on feedlot performance: a review. J Anim Sci. 1998;76(1):272–274. | ||
Wittum TE, Woollen NE, Perino LJ, Littledike ET. Relationships among treatment for respiratory tract disease, pulmonary lesions evident at slaughter, and rate of weight gain in feedlot cattle. J Am Vet Med Assoc. 1996;209(4):814–818. | ||
Rajala-Schultz PJ, Torres AH, Degraves FJ, Gebreyes WA, Patchanee P. Antimicrobial resistance and genotypic characterization of coagulase-negative staphylococci over the dry period. Vet Microbiol. 2009;134(1–2):55–64. | ||
Johnson KK, Pendell DL. Market impacts of reducing the prevalence of bovine respiratory disease in United States beef cattle feedlots. Front Vet Sci. 2017;4:189. | ||
Headley SA, Okano W, Balbo LC, et al. Molecular survey of infectious agents associated with bovine respiratory disease in a beef cattle feedlot in southern Brazil. J Vet Diagn Invest. 2017;30(2):249–251. | ||
Holman DB, Klima CL, Ralston BJ, et al. Metagenomic sequencing of bronchoalveolar lavage samples from feedlot cattle mortalities associated with bovine respiratory disease. Genome Announc. 2017;5(40):e01045-17. | ||
Gagea MI, Bateman KG, van Dreumel T, et al. Diseases and pathogens associated with mortality in Ontario beef feedlots. J Vet Diagn Invest. 2006;18(1):18–28. | ||
Haines DM, Martin KM, Clark EG, Jim GK, Janzen ED. The immunohistochemical detection of Mycoplasma bovis and bovine viral diarrhea virus in tissues of feedlot cattle with chronic, unresponsive respiratory disease and/or arthritis. Can Vet J. 2001;42(11):857. | ||
Jensen R, Pierson R, Braddy P, et al. Shipping fever pneumonia in yearling feedlot cattle. J Am Vet Med Assoc. 1976;169(5):500–506. | ||
Shahriar FM, Clark EG, Janzen E, West K, Wobeser G. Coinfection with bovine viral diarrhea virus and Mycoplasma bovis in feedlot cattle with chronic pneumonia. Can Vet J. 2002;43(11):863. | ||
Welsh RD, Dye LB, Payton ME, Confer AW. Isolation and antimicrobial susceptibilities of bacterial pathogens from bovine pneumonia: 1994–2002. J Vet Diagn Invest. 2004;16(5):426–431. | ||
Hoerlein A, Marsh C. Studies on the epizootiology of shipping fever in calves. J Am Vet Med Assoc. 1957;131(3):123–127. | ||
Maday J. Back to basics with BRD. Canadian Cattlemen the Beef Magazine. 2014 Dec: 1–4. | ||
Apley M. Bovine respiratory disease: pathogenesis, clinical signs, and treatment in lightweight calves. Vet Clin North Am Food Anim Pract. 2006;22(2):399–411. | ||
Ellis JA. The immunology of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract. 2001;17(3):535–550, vi–vii. | ||
Turni C, Dayao D, Aduriz G, et al. A Pasteurella multocida strain affecting nulliparous heifers and calves in different ways. Vet Microbiol. 2016;195:17–21. | ||
Blum S, Freed M, Zukin N, et al. Bovine subclinical mastitis caused by Mannheimia granulomatis. J Vet Diagn Invest. 2010;22(6):995–999. | ||
Larson RL, Step DL. Evidence-based effectiveness of vaccination against Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni in feedlot cattle for mitigating the incidence and effect of bovine respiratory disease complex. Vet Clin North Am Food Anim Pract. 2012;28(1):97–106, 106e101–107, ix. | ||
Checkley SL, Campbell JR, Chirino-Trejo M, Janzen ED, Waldner CL. Associations between antimicrobial use and the prevalence of antimicrobial resistance in fecal Escherichia coli from feedlot cattle in western Canada. Can Vet J. 2010;51(8):853–861. | ||
Lindsay MA. Target discovery. Nat Rev Drug Discov. 2003;2(10):831–838. | ||
Windeyer C, Timsit E, Barkema H. Bovine respiratory disease in pre-weaned dairy calves: are current preventative strategies good enough? Vet J. 2017;224:16–17. | ||
Perumal D, Lim CS, Sakharkar KR, Sakharkar MK. Differential genome analyses of metabolic enzymes in Pseudomonas aeruginosa for drug target identification. In Silico Biol. 2007;7(4, 5):453–465. | ||
Sakharkar KR, Sakharkar MK, Chow VT. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. In Silico Biol. 2004;4(3):355–360. | ||
Hossain MU, Khan MA, Hashem A, et al. Finding potential therapeutic targets against shigella flexneri through proteome exploration. Front Microbiol. 2016;7:1817. | ||
Hadizadeh M, Tabatabaiepour SN, Tabatabaiepour SZ, Hosseini Nave H, Mohammadi M, Sohrabi SM. Genome-wide identification of potential drug target in enterobacteriaceae family: a homology-based method. Microb Drug Resist. 2017;24(1):8–17. | ||
Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci U S A. 1996;93(19):10268–10273. | ||
Glass JI, Assad-Garcia N, Alperovich N, et al. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A. 2006;103(2):425–430. | ||
Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12(6):962–968. | ||
Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23(11):2072–2080. | ||
Wadood A, Jamal A, Riaz M, et al. Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb Pathog. 2017;115:194–198. | ||
Uddin R, Siddiqui QN, Azam SS, Saima B, Wadood A. Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant Mycobacterium tuberculosis (XDR KZN 605) through subtractive genomics approach. Eur J Pharm Sci. 2018;114:13–23. | ||
Jamal SB, Hassan SS, Tiwari S, et al. An integrative in-silico approach for therapeutic target identification in the human pathogen Corynebacterium diphtheriae. PLoS One. 2017;12(10):e0186401. | ||
Birhanu BT, Lee SJ, Park NH, Song JB, Park SC. In silico analysis of putative drug and vaccine target of the metabolic pathways of Actinobacillus pleuropneumoniae using subtractive/comparative genomics approach. J Vet Sci. 2017;19(2):188–199. | ||
Hossain T, Kamruzzaman M, Choudhury TZ, Mahmood HN, Nabi A, Hosen MI. Application of the subtractive genomics and molecular docking analysis for the identification of novel putative drug targets against Salmonella enterica subsp. enterica serovar Poona. BioMed Res Int. 2017;2017:3783714. | ||
Agüero F, Al-Lazikani B, Aslett M, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7(11):900–907. | ||
Luo H, Lin Y, Gao F, Zhang C-T, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2013;42(D1):D574–D580. | ||
Singh S, Singh DB, Singh A, et al. An approach for identification of novel drug targets in Streptococcus pyogenes SF370 through pathway analysis. Interdiscip Sci. 2016;8(4):388–394. | ||
Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. | ||
Wishart DS, Wu A. Using DrugBank for in silico drug exploration and discovery. Curr Protoc Bioinformatics. 2016;54:14.4.1–14.4.31. | ||
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35 (Suppl 2):W182–W185. | ||
Caspi R, Altman T, Billington R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42(Database issue):D459–D471. | ||
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. | ||
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. | ||
Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44(D1):D694–D697. | ||
Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25(21):2857–2859. | ||
Kim AB. Virulence Mechanisms of Bacterial Pathogens. 3rd ed. New Delhi, India: 2000. | ||
Dickey SW, Cheung GYC, Otto M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov. 2017;16(7):457–471. | ||
Peng C, Gao F. Protein localization analysis of essential genes in prokaryotes. Sci Rep. 2014;4:6001. | ||
Acharya A, Garg LC. Drug target identification and prioritization for treatment of ovine foot rot: an In Silico approach. Int J Genomics. 2016;2016:7361361. | ||
Duff GC, Galyean ML. Board-invited review: recent advances in management of highly stressed, newly received feedlot cattle. J Anim Sci. 2007;85(3):823–840. | ||
Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. The epidemiology of bovine respiratory disease: what is the evidence for predisposing factors? Can Vet J. 2010;51(10):1095–1102. | ||
McVey DS. BRD research needs in the next 10–20 years. Anim Health Res Rev. 2009;10(2):165–167. | ||
Urban-Chmiel R, Grooms D. Prevention and control of bovine respiratory disease. J Livestock Sci. 2012;3:27–36. | ||
Johnston D, Earley B, Cormican P, et al. Illumina MiSeq 16S amplicon sequence analysis of bovine respiratory disease associated bacteria in lung and mediastinal lymph node tissue. BMC Vet Res. 2017;13(1):118. | ||
Van Donkersgoed J. Meta-analysis of field trials of antimicrobial mass medication for prophylaxis of bovine respiratory disease in feedlot cattle. Can Vet J. 1992;33(12):786. | ||
Cameron A, McAllister TA. Antimicrobial usage and resistance in beef production. J Anim Sci Biotechnol. 2016;7(1):68. | ||
Cusack PM. Effect of mass medication with antibiotics at feedlot entry on the health and growth rate of cattle destined for the Australian domestic market. Aust Vet J. 2004;82(3):154–156. | ||
Frank GH, Briggs RE, Duff GC, Loan RW, Purdy CW. Effects of vaccination prior to transit and administration of florfenicol at time of arrival in a feedlot on the health of transported calves and detection of Mannheimia haemolytica in nasal secretions. Am J Vet Res. 2002;63(2):251–256. | ||
Step DL, Engelken T, Romano C, et al. Evaluation of three antimicrobial regimens used as metaphylaxis in stocker calves at high risk of developing bovine respiratory disease. Vet Ther. 2007;8(2):136–147. | ||
Wellman NG, O’Connor AM. Meta-analysis of treatment of cattle with bovine respiratory disease with tulathromycin. J Vet Pharmacol Ther. 2007;30(3):234–241. | ||
Guthrie C, Rogers K, Christmas R, Vogel G, Laudert S, Mechor G. Efficacy of metaphylactic tilmicosin for controlling bovine respiratory disease in high-risk northern feeder calves. Bovine Practitioner. 2004:46–53. | ||
Owens J. Determining druggability. Nat Rev Drug Discov. 2007;6(3):187. | ||
Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14(5):917–924. | ||
Catry B, Dewulf J, Maes D, et al. Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS One. 2016;11(1):e0146488. | ||
Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–316. | ||
Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–715. | ||
Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40–51. | ||
Corsello SM, Bittker JA, Liu Z, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017;23(4):405–408. | ||
Knoben JE, Scott GR, Tonelli RJ. An overview of the FDA publication approved drug products with therapeutic equivalence evaluations. Am J Hosp Pharm. 1990;47(12):2696–2700. | ||
Lees P, Potter T, Pelligand L, Toutain PL. Pharmacokinetic-pharmacodynamic integration and modelling of oxytetracycline for the calf pathogens Mannheimia haemolytica and Pasteurella multocida. J Vet Pharmacol Ther. 2018;41(1):28–38. | ||
Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt Sinai J Med. 2007;74(1):27–32. | ||
Juhas M, Eberl L, Church GM. Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol. 2012;30(11):601–607. | ||
Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21(10):562–568. | ||
Suthers PF, Zomorrodi A, Maranas CD. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol Syst Biol. 2009;5:301. | ||
Tucker CL, Fields S. Lethal combinations. Nat Genet. 2003;35(3):204–205. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




