Back to Journals » Drug Design, Development and Therapy » Volume 9
HDACiDB: a database for histone deacetylase inhibitors
Authors Murugan K , Sangeetha S, Ranjitha S, Vimala AD, Al-Sohaibani S, Rameshkumar G
Received 27 November 2014
Accepted for publication 10 February 2015
Published 20 April 2015 Volume 2015:9 Pages 2257—2264
DOI https://doi.org/10.2147/DDDT.S78276
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Shu-Feng Zhou
Kasi Murugan,1 Shanmugasamy Sangeetha,2 Shanmugasamy Ranjitha,2 Antony Vimala,2 Saleh Al-Sohaibani,1 Gopal Rameshkumar2
1Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia; 2Bioinformatics Laboratory, Anna University K. Balachander Research Centre, MIT Campus of Anna University Chennai, Chennai, India
Abstract: An histone deacetylase (HDAC) inhibitor database (HDACiDB) was constructed to enable rapid access to data relevant to the development of epigenetic modulators (HDAC inhibitors [HDACi]), helping bring precision cancer medicine a step closer. Thousands of HDACi targeting HDACs are in various stages of development and are being tested in clinical trials as monotherapy and in combination with other cancer agents. Despite the abundance of HDACi, information resources are limited. Tools for in silico experiments on specific HDACi prediction, for designing and analyzing the generated data, as well as custom-made specific tools and interactive databases, are needed. We have developed an HDACiDB that is a composite collection of HDACi and currently comprises 1,445 chemical compounds, including 419 natural and 1,026 synthetic ones having the potential to inhibit histone deacetylation. Most importantly, it will allow application of Lipinski’s rule of five drug-likeness and other physicochemical property-based screening of the inhibitors. It also provides easy access to information on their source of origin, molecular properties, drug likeness, as well as bioavailability with relevant references cited. Being the first comprehensive database on HDACi that contains all known natural and synthetic HDACi, the HDACiDB may help to improve our knowledge concerning the mechanisms of actions of available HDACi and enable us to selectively target individual HDAC isoforms and establish a new paradigm for intelligent epigenetic cancer drug design. The database is freely available on the http://hdacidb.bioinfo.au-kbc.org.in/hdacidb/ website.
Keywords: cancer, drug likeness, histone deacetylase inhibitors, epigenetics, Lipinski’s rule, molecular properties
Introduction
Cancer is the second leading cause of death in humans. Today, wide-ranging cancer-oriented sequencing efforts are yielding new insights not only into the disease biology of cancer but also into the development of pathway-driven targeted therapy. The value of such approaches seems undeniable.1 The World Health Organization’s International Agency for Research on Cancer online database, GLOBOCAN 2012, estimated that there were over 14.1 million new cancer cases and 8.2 million cancer-related deaths in 2012, compared with about 12.7 million and 7.6 million, respectively, in 2008. It also projects a substantial increase to 19.3 million new cancer cases by 2025, as a result of growth and aging of the global population.2 According to the National Cancer Institute estimation, by 2020 cancer-related medical costs will reach not less than US$158 billion.3
Cancer involves the uncontrolled growth and abnormal spread of cells and is now known as a genetic and epigenetic abnormality-associated disease. Global changes in the chromatin structure of cancer cells lead to alterations in nuclear functions, including gene expression. Among the regulatory epigenetic mechanisms, histone protein modification has been well studied.4 The potentially reversible nature of epigenetic modifications, such as histone deacetylase (HDAC)-mediated acetyl group removal from the backbone of histones, leads to chromatin compaction and, ultimately, transcriptional repression.5 The epigenetic modifications ability to induce heritable silencing of genes without a change in their coding sequence6 makes them attractive targets for cancer treatment and anticancer drug development.
An 18-gene family encodes these HDAC proteins, and based on their sequence homology and domain organization, the family has been grouped into four classes. The typical class I HDACs that are localized in the nucleus include HDAC1, HDAC2, HDAC3, and HDAC8. Class II HDACs are made up of proteins that reside in both the nucleus and cytoplasm (class IIa, HDAC4, HDAC5, HDAC7, and HDAC9; class IIb, HDAC6 and HDAC10). HDAC11, localized in the nucleus, is the only HDAC member of class IV. All of these enzymes function by using a metal ion-dependent mechanism. Class III HDACs consist of yeast homolog NAD+-dependent sirtuins (SIRTs 1–7), showing function-dependent localization in various organelles.7–9 Overexpression, increased activity, and appearance of different isoforms of HDACs play a significant role in carcinogenesis and tumor progression. Thus, HDAC inhibitors (HDACi) have garnered worldwide attention as promising agents for epigenetic cancer therapy.7
Recent advances in HDAC protein recombinant expression and purification have permitted characterization of their inhibitory profile. Compounds targeting “classical” class I, II, and IV HDACs and those in clinical trial evaluation are commonly called HDACi.10 The varied large collection of HDACi molecules, initially developed as single anticancer agents, is now cataloged as epigenetic modulators, because they have the ability to modulate gene expression through numerous direct and indirect ways.11 Their activity results in cell-cycle arrest, mitotic catastrophe, and programmed cell death, and causes angiogenesis-like antitumor effects.8 Their gene-modulating chemosensitivity, immunogenicity-enhancing activity, or cancer cell innate antiviral response-reducing activity in combination with chemotherapy, immunotherapy, or oncolytic virotherapy have shown better efficacy.8
Currently, in preclinical studies, potent and specific anticancer activity has been recognized among a number of classes of HDACi.5 First-generation drugs such as suberoylanilide hydroxamic acid (vorinostat; Zolinza™, Merck, Whitehouse Station, TX, USA) and romidepsin (FK228; Istodax™, Celgene, Summit, NJ, USA) have already been approved by the US Food and Drug Administration in 2006 and 2009, respectively, for the treatment of cutaneous T-cell lymphomas. Notwithstanding their approval for clinical use, their mechanism of action as anticancer agents has not been completely elucidated.12 Hence, the actual clinical responses to these HDACi have been less consistent and weaker than expected. A number of side effects have also been observed in several cases.
All these issues have been attributed to their lack of HDAC type-specific inhibition.5 Because HDACi modify expression of many genes, inhibition of one isoform may result in therapeutically beneficial epigenetic changes, while inhibition of another isoform could cause harmful or unwanted changes. Selective HDACi circumvent these types of situations and show less toxicity than pan-HDACi.13 Designing potential type-specific HDACi compounds remains an enduring research interest in cancer drug development. Hence, “the knowledge and understanding of the various structural variants of the HDACis and their pharmacophore properties can be useful in the rational design and development of more potent isoform selective HDACis”.14 Addition of certain structural moieties could result in isoform-selective HDACi.13 Available X-ray crystal and three-dimensional structures of the HDAC enzyme active site and information on their proposed catalytic function will allow an in silico approach for the initial screening and designing of HDAC isoform-selective HDACi at a reduced cost.
Hence, we believe a database housing a rapidly increasing list of HDACi as well as elucidation of their exact mechanism of action, along with a knowledge of structural elements responsible for activity, would ease the identification or designing of new selective HDACi with HDAC class-specific inhibition. The relational HDACi database (HDACiDB) has been developed with the aim of providing relevant information about all natural and synthetic HDACi currently in clinical trials. It also provides the necessary drug development screening information, including physicochemical properties, two-dimensional and three-dimensional structures with retrieval links, and biological potency, safety, and pharmacokinetics for enabling easy elucidation of their pharmacokinetic and toxicological properties with relevant literature sources. This database not only helps to expand our knowledge on HDACi but also enables rapid access to allow in silico screening of novel HDACi and developmental studies.
Description of the HDACiDB database
The HDACiDB (http://hdacidb.bioinfo.au-kbc.org.in/hdacidb/) is organized and constructed around publicly available literature and is based on HDACi main compounds collected from open literature, patent databases, and clinical trial compound databases approved by the US Food and Drug Administration. The main purpose of this HDACiDB platform is to provide a comprehensive resource to organize and present all HDAC-targeted drug development screening information in a convenient and straightforward way. It mainly allows screening of HDACi based on Lipinski’s rule of five as well as other physicochemical parameters. The Lipinski’s rule of five describes the ADME (absorption, distribution, metabolism, and excretion) properties of a drug that help in evaluating drug likeness and bioavailability. The database is built using the Apache HyperText Transfer Protocol web server along with the MySQL database server and is available as an HTML document. A schematic representation of the HDACiDB, showing the homepage, search options (name, property, Lipinski’s rule, and other drug likeness parameters) available separately for natural and synthetic compounds, the developer team, contact information, and more is shown in Figure 1.
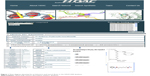  | Figure 1 Flow diagram showing the architecture and search flows in the HDACiDB database. |
Data collection
The relevant literature data were collected using search engines such as PubMed, Web of Science, and Google and were used for input loading of information into the database. Search terms such as HDAC, HDACi, Lipinski’s rule, drug likeness, and bioavailability were each used one at a time. Terms such as epigenetic drugs, phytochemicals, and chemoprevention were used in conjunction with the Boolean operation. The collected literature was dealt with manually, and all resultant information inputs were loaded into the database. Structural and experimental data for main compounds were collected manually from the NCBI-PubChem15 and DrugBank databases.16 Similarly, analog compounds having the same structural backbone but with different side chains were collected from the ChemBank database using a similarity search.17 Structures of collected compounds were drawn using conventional ChemSketch version 12.0 software18 and saved as a file. Before importing these structures into the database, they were manually checked for atom valence and correctness of representation. To augment the expansion of the available data, a data submission page has been put up on the website, allowing moderators to see new inputs from researchers working on HDACi.
Chemical and molecular properties
For compounds in the database, the International Union of Pure and Applied Chemistry (IUPAC) name, PubChem/ChemBank identifier, molecular formula, canonical SMILES (Simplified Molecular-Input Line-Entry System) information, international chemical identifier (InCHI), tautomer count, exact mass, as well as monoisotropic mass were collected from the PubChem, DrugBank, and ChemBank databases.15–17 Their molecular properties, including membrane permeability and oral bioavailability, were calculated using the basic molecular descriptors, such as partition coefficient (LogP), total polar surface area, molecular weight, number of atoms (natoms), number of violations (nviolations), number of rotatable bonds (nrotb), number of hydrogen bond donors, and number of hydrogen bond acceptors. The protease inhibitor, G protein-coupled receptor ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, enzyme inhibitor, and volume scores indicating the drug likeness were also calculated. These drug likeness scores allow efficient separation of active and inactive molecules based on their similarity to drug’s already in use or under clinical trials molecular properties and structural features. The online Molinspiration cheminformatics software was used for these calculations.19
Data management
The HDACiDB has a flexible and open design that can accommodate a variety of inputs on HDACi from both curators and other submitters. The HDACiDB was developed using common all-purpose scripting language hypertext preprocessor functions.20 Tasks were written to analyze, extract, reformat, and construct data elements, and built storage was constructed using the MySQL21 database management system for storage and for querying all of the data. External file data were stored in the SQL database in a single major table format. Each compound was stored in an SQL table with a unique HDACi identifier. The HDACi identifier of a compound was assigned in such a way as to indicate its origin, parent, or similar compound. For example, for CHAn000, the first three capital letters represent the compound name chlamydocin. The small letter “n” represents its origin as a natural compound (“s” if it is of synthetic origin), and assigned numbers represent whether it belongs to the parent compound or similar compound. Here, the number “000” represents a parent compound, whereas “001” would represent a similar compound-1. These data were processed using jQuery Slider, and the JMOL applet (http://www.jmol.org) was employed for the visualization of two-dimensional and three-dimensional compound structures.
At present, the HDACiDB contains a total of 1,445 HDAC inhibitors, with 419 natural compounds and 1,026 synthetic compounds. It includes 33 natural parent compounds and 386 analog compounds (Table S1), and 25 synthetic parent compounds and 1,001 analog compounds (Table S2).
Database features
The HDACiDB holds the potential for users to conduct structure-activity relationship-based facile searches for novel lead compounds as potential specific HDACi using Lipinski’s rule of five, a set of simple molecular descriptors describing overall drug likeness and other physicochemical properties. Lipinski’s rule of five states that most “drug-like” molecules must match the following criteria: LogP ≤5, molecular weight ≤500, number of hydrogen bond acceptors ≤10 (n OH), and number of hydrogen bond donors ≤5 (n NH or OH). It also allows virtual screening of suitable HDACi using other properties, including total polar surface area, G protein-coupled receptor ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, enzyme inhibitor, natoms, nviolations, nrotb, and protease inhibitor. The search requires users to initialize the value for each property within which compounds fall. Using the interactive scrollers, the initializing value can be done. Running the search will return a set of compounds that obey the given parameters.
Querying and HDACiDB results display
HDACi compounds can be viewed and retrieved from the HDACiDB. The HDACiDB offers a search engine to query through two distinct (natural and synthetic) entry points for HDACi compound searching. Both natural and synthetic compound data can be accessed separately by either their “name” or their “property” track. Figure 2 provides the information flow during a compound search. Search by name allows the user to select a main compound and view the group of similar compounds available in the list. Segregation of properties into mainly two types, namely drug properties and physicochemical properties, allows the user to perform compound searches by applying the desired Lipinski’s rule or using other properties.
Drug property information comprises the IUPAC name of the compound, the PubChem/ChemBank identifier, HDACiDB identifier, molecular formula, canonical SMILES, InCHI, tautomer count, exact mass, and monoisotopic mass. The compound LogP, total polar surface area, molecular weight, natoms, nviolations, nrotb, hydrogen acceptor, number of hydrogen bond donors, protease inhibitor, G protein-coupled receptor ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, enzyme inhibitor, and volume are the incorporated physicochemical properties. Hence, the provided Lipinski’s rule parameters (molecular weight, LogP, number of hydrogen bond donors, and number of hydrogen bond acceptors) sliders allow for the selection of values and search initialization. The same provision is also available for selecting compounds based on other drug likeness parameters. After submission of the compound’s first alphabet, all the compounds beginning with that letter appear, and selection of the particular compound will lead to the display of a page having compound and related information. The compound IUPAC name, its main compound, physicochemical and drug properties, compound group, two-dimensional as well as three-dimensional structure, and its description are all displayed.
To get more details about each feature, the user can click on the particular link and a new page will appear to display the queried information. “Physicochemical” will give LogP, total polar surface area, molecular weight, its number of atoms the search molecule contains, OHNH, ON, Lipinski’s rule violation, rotatable bond, and volume. It also gives its G protein-coupled receptor ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, and protease inhibitor, as well as enzyme inhibitor interactive docking scores. “Drug properties” include PubChem identifier, molecular formula, canonical SMILES, InCHI, tautomer count, exact mass, and monoisotopic mass. “Compound group” yields the group to which it belongs and the other compounds of this group. “All properties” delivers a compilation of all these properties in a single page. “Compound description” provides a description of the compound, including its current pharmaceutical status and other applications. “Literature link” provides updates of the current status of the particular inhibitor and all publically available literature. “Export molecular file” allows the export and downloading of a compound’s molecular file. Views of the two-dimensional structure and Jmol three-dimensional structure of the compounds are also available. Hence, searching the HDACiDB would allow a structure-activity relationship-based facile selection of lead HDACi compounds that are specific for one of the non-redundant HDACs.
Future development
In light of the fact that information on these epigenetic modulators continues to grow with inputs from precision cancer medicine development programs, preclinical studies, and other research, this new information would be incorporated into the HDACiDB on a regular basis. A major focus will be to extend the database through acquisition of more experimental data and to add a number of Java plug-ins to facilitate virtual screening as well as to enrich the HDACiDB with more features to make it more interactive and user-friendly. This database serves as a foundation and launching point for further studies on HDACi and their analog compounds, and as an enhancer of the success rate of computer-aided cancer drug design. In future, all entries would be hyperlinked to other major cancer-related databases to provide information beyond the scope of the HDACiDB.
Acknowledgments
This project was supported by King Saud University, Deanship of Scientific Research, College of Sciences Research center.
Disclosure
The authors report no conflicts of interest in this work.
References
Steensma DP. The beginning of the end of the beginning in cancer genomics. N Engl J Med. 2013;368(22):2138–2140. | ||
World Health Organization. Latest world cancer statistics. 2013. Available from: http://www.iarc.fr/en/media-centre/pr/2013/pdfs/pr223_E.pdf. Accessed October 10, 2014. | ||
Mariotto AB, Yabroff KR, Shao Y, et al. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103(2):117–128. | ||
Koturbash I, Simpson NE, Beland FA, Pogribny IP. Alterations in histone 4 lysine 20 methylation: implications for cancer detection and prevention. Antioxid Redox Signal. 2012;17(2):365–374. | ||
Botrugnoa OA, Santoroa F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280(2):134–144. | ||
Monneret C. Histone deacetylase inhibitors for epigenetic therapy of cancer. Anticancer Drugs. 2007;18(4):363–370. | ||
Di MS, Chini MG, Terracciano S, Bruno I, Riccio R, Bifulco G. Structural basis for the design and synthesis of selective HDAC inhibitors. Bioorg Med Chem. 2013;21(13):3795–3807. | ||
Yu X, Guo ZS. Epigenetic drugs for cancer treatment and prevention: mechanisms of action. Biomol Concepts. 2010;1(3–4):239–251. | ||
Sangeetha S, Ranjitha S, Murugan K, Rameshkumar G. Breast cancer specific HDACi lead discovery using molecular docking and descriptor study. Trends in Bioinformatics. 2013;6(2):25–44. | ||
Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. | ||
Bertrand P. Inside HDAC with HDAC inhibitors. Eur J Med Chem. 2010;45(6):2095–2116. | ||
Sweet MJ, Shakespear MR, Kamal NA, Fairlie DP. HDAC inhibitors: modulating leukocyte differentiation, survival, proliferation and inflammation. Immunol Cell Biol. 2012;90(1):14–22. | ||
Kozikowski AP, Butler KV. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14(6):505–528. | ||
Kalyaanamoorthy S, Chen YPP. Energy based pharmacophore mapping of HDAC inhibitors against class I HDAC enzymes. Biochim Biophys Acta. 2013;1834(1):317–328. | ||
Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15(23–24):1052–1057. | ||
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(database issue):D901–D906. | ||
Seiler KP, George GA, Happ MP, et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. | ||
Osterberg T, Norinder U. Prediction of drug transport processes using simple parameters and PLS statistics: the use of ACD/logP and ACD/ChemSketch descriptors. Eur J Pharm Sci. 2001;12(3):327–337. | ||
AliAsghar J, Jihane F, Mostafa M, et al. Petra, Osiris and Molinspiration (POM) together as a successful support in drug design: antibacterial activity and biopharmaceutical characterization of some azo Schiff bases. Med Chem Res. 2012;21(8):1984–1990. | ||
Xie SX, Baek Y, Grossman M, et al. Building an integrated neurodegenerative disease database at an academic health center. Alzheimers Dement. 2011;7(4):84–93. | ||
Sharman JL, Gerloff DL. MaGnET: Malaria genome exploration tool. Bioinformatics. 2013;29(18):2350–2352. |
Supplementary materials
  | Table S1 Details of the content of the histone deacetylase inhibitor database on parent natural the original main compounds |
  | Table S2 Details of the content of the histone deacetylase inhibitor database for the main original synthetic compounds |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

