Back to Journals » International Journal of Nanomedicine » Volume 15
Fullerene Derivatives as Lung Cancer Cell Inhibitors: Investigation of Potential Descriptors Using QSAR Approaches
Authors Huang HJ, Kraevaya OA , Voronov II, Troshin PA, Hsu S
Received 23 December 2019
Accepted for publication 3 March 2020
Published 14 April 2020 Volume 2020:15 Pages 2485—2499
DOI https://doi.org/10.2147/IJN.S243463
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yan Shen
Hung-Jin Huang,1,2 Olga A Kraevaya,3,4 Ilya I Voronov,4 Pavel A Troshin,3,4 Shan-hui Hsu1,2,5
1Institute of Polymer Science and Engineering, National Taiwan University, Taipei, Taiwan; 2Institute of Cellular and System Medicine, National Health Research Institutes, Miaoli, Taiwan; 3Skolkovo Institute of Science and Technology, Moscow, Russian Federation; 4Institute for Problems of Chemical Physics of Russian Academy of Sciences, Chernogolovka, Russian Federation; 5Research and Development Center for Medical Devices, National Taiwan University, Taipei, Taiwan
Correspondence: Shan-hui Hsu
Institute of Polymer Science and Engineering, National Taiwan University, No. 1, Sec. 4 Roosevelt Road, Taipei 10617, Taiwan
Tel +886-2-3366-5313
Fax +886-2-3366-5237
Email [email protected]
Pavel A Troshin
Institute for Problems of Chemical Physics of Russian Academy of Sciences, Chernogolovka 142432, Russian Federation
Tel +7 496522-1418
Fax +7 496515-5420
Email [email protected]
Background: Nanotechnology-based strategies in the treatment of cancer have potential advantages because of the favorable delivery of nanoparticles into tumors through porous vasculature.
Materials and Methods: In the current study, we synthesized a series of water-soluble fullerene derivatives and observed their anti-tumor effects on human lung carcinoma A549 cell lines. The quantitative structure–activity relationship (QSAR) modeling was employed to investigate the relationship between anticancer effects and descriptors relevant to peculiarities of molecular structures of fullerene derivatives.
Results: In the QSAR regression model, the evaluation results revealed that the determination coefficient r2 and leave-one-out cross-validation q2 for the recommended QSAR model were 0.9966 and 0.9246, respectively, indicating the reliability of the results. The molecular modeling showed that the lack of chlorine atom and a lower number of aliphatic single bonds in saturated hydrocarbon chains may be positively correlated with the lung cancer cytotoxicity of fullerene derivatives. Synthesized water-soluble fullerene derivatives have potential functional groups to inhibit the proliferation of lung cancer cells.
Conclusion: The guidelines obtained from the QSAR model might strongly facilitate the rational design of potential fullerene-based drug candidates for lung cancer therapy in the future.
Keywords: water-soluble fullerene derivatives, non-small cell lung cancer, cytotoxicity, machine learning, QSAR
Background
Lung cancer is one of the leading causes of death around the world.1 Lung cancer can be classified into two main types, non-small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC).2 Approximately 85% of patients with lung cancer are diagnosed with NSCLC, and the survival rate remains below 15%.3,4 Lung cancer cells can metastasize to bones, brain, and adrenal glands via blood vessels and lymphatic routes.5 Surgery is not a suitable treatment for the most lung cancer patients,6 while chemotherapy has become a benefit strategy.7 To date, the mechanisms of lung cancer metastasis are not fully understood. Clinically, chemotherapy is often used to treat the metastatic lung cancer in combination with surgery and radiotherapy.8,9 However, the efficacy of the existing chemotherapeutics for lung cancer is limited by several drawbacks such as insufficient drug concentrations in tumors, drug resistance of tumor cells, and systemic toxicity. Recent attention has been paid to the development of more selective anticancer agents to treat the cancer and to minimize the side effects.10–13 The characteristic median lethal dose IC50 of a certain compound acting on cancer cells and suppressing their proliferation by 50% is usually considered as a figure of merit in initial high-throughput screening of potential drug candidates in cell-based assays.10
Nanotechnology has been widely used in biomedicine for the development of drug delivery systems, biological labels, protein detection, tumor destruction, etc.14 Tumors have immature and porous vasculature that provides an access for nanoparticles to enter the circulation system,15 so nanoparticles can be used for selective delivery of drugs to the cancer cells. Carbon-based nanoparticles are considered as very promising and highly biocompatible drug carriers. One of the nanocarbon forms, fullerenes, has been first experimentally observed by Kroto et al in 1985.16 A unique combination of chemical, physical and electronic properties of fullerenes makes them an interesting family of materials for a variety of applications including their use in biomedicine. However, fullerenes are highly hydrophobic and poorly soluble in aqueous medium. For drug development, attachment of solubilizing polar groups to the carbon cage has been used to modify fullerenes and overcome the issue of solubility in physiological media. Water-soluble fullerene derivatives with various types of biological activity have been synthesized and their potential therapeutic use for certain diseases has been featured.17,18 Previously, we observed that water-soluble fullerene derivatives could protect brain neural stem cells or kill brain tumor cells, depending on their surface functional groups.19 However, identifying the chemical structure of addends and their optimal arrangement on the carbon cage enabling the anticancer effect represents still a big challenge.
Computer-aided drug design (CADD) is an efficient tool to provide the rapid identification of potent candidates targeting specific diseases, which has been used to optimize the design of therapeutic compounds. The CADD technique can be divided into two general categories: structure-based drug design and ligand-based drug design.20 The structure-based CADD is implemented to design of compounds based on the knowledge of the target protein structure, while the ligand-based CADD can be utilized to develop the relationship between molecular structures and biological activities. The quantitative structure–activity relationship (QSAR) is one of ligand-based CADD techniques employed in molecular biology and medicinal chemistry, which is also called as indirect drug design. The QSAR can provide predictive models based on mathematical and statistical relations that can be used to optimize the design of known types of chemical drugs to achieve an improved activity.21,22 However, there are still very few studies focused on the design of nanomedicines with the assistance of QSAR or other CADD techniques.23 Here, we explored the potential of developing predictive computer models for a series of water-soluble fullerene derivatives acting as inhibitors of NSCLC cancer cells. Indeed, we succeeded in building a QSAR model considering the peculiarities of the chemical structures of water-soluble fullerene derivatives and the experimental IC50 values reflecting their anticancer activity. We showed that the information offered by this model might guide the design of water-soluble fullerene derivatives with improved efficacy when used as anticancer therapeutics.
Methods
Synthesis of Water-Soluble Fullerene Derivatives
Fullerene derivatives Cs-C60Ar6, C60Ar5X (X=Cl, H) and Cs-C60Ar4 (compounds 1, 3–10) were synthesized using reported approach based on the arylation of the chlorofullerene C60Cl6 with the esters of aromatic acids followed by acidic hydrolysis of ester groups (Figure 1).24 Preparative chromatography was used to separate untypical products Cs-C60Ar6 and Cs-C60Ar4. Compound 2 with the structure C60Ar5Et was obtained by treatment of the fullerene derivative 1 with P(OEt)3 at 100°C as reported previously.25 Compounds 3, 4 and 8 were synthesized and fully characterized earlier.26 Compounds 1, 2, 5, 6, 7, 9, 10 were obtained here for the first time (see Figures S1–S37 for NMR and ESI MS data).
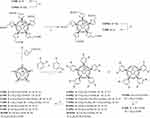 |
Figure 1 Synthesis of the fullerene derivatives used in this work. Conditions: i – FeCl3, PhNO2, 100oC, 1h; ii – P(OEt)3, PhCH3, 100oC, 1h; iii – HCl, CH3COOH, PhCH3, 70oC, 3d. |
Compound 1-OMe. (79%)1H NMR (500 MHz, CDCl3, δ, ppm): 3.71 (s, 3H), 3.74 (s, 6H), 3.76 (s, 6H), 3.78 (s, 2H), 3.83 (s, 4H), 3.87 (s, 4H), 6.81 (d, 1H, J = 3.6 Hz), 6.85–6.87 (m, 3H), 6.91 (d, 2H, J = 3.5 Hz), 7.10 (d, 2H, J = 3.5 Hz), 7.41 (d, 2H, J = 3.6 Hz).13C NMR (125 MHz, CDCl3, δ, ppm): 35.38 (CH2), 35.49 (CH2), 35.62 (CH2), 52.29 (CH3), 52.32 (CH3), 52.34 (CH3), 54.03 (Csp3 fullerene cage), 56.19 (Csp3 fullerene cage), 59.60 (Csp3 fullerene cage), 75.30 (Csp3 fullerene cage-Cl), 126.48, 126.92, 126.95, 127.58, 129.79, 135.68, 135.93, 136.24, 140.52, 141.56, 142.14, 142.59, 142.83, 143.17, 143.46, 144.07, 144.29, 144.34, 144.59, 144.69, 144.97, 145.78, 146.66, 147.17, 147.33, 147.85, 148.21, 148.32, 148.37, 148.69, 148.76, 149.67, 150.40, 153.19, 155.55, 170.60 (COOCH3), 170.64 (COOCH3), 170.71 (COOCH3). FTIR (KBr pellet, ν, cm−1): 538 (M), 754 (M), 778 (M), 798 (M), 1000 (M), 1038 (M), 1166 (S), 1212 (S), 1262 (M), 1288 (M), 1310 (M), 1348 (M), 1404 (M), 1432 (M), 1460 (M), 1542 (M), 1560 (M), 1654 (M), 1736 (VS), 2336 (M), 2586 (M), 2850 (M), 2922 (M), 3396 (M), 3406 (M), 3448 (M), 3506 (W).
Compound 1-OH. (97%)1H NMR (500 MHz, (CD3)2SO, δ, ppm): 3.75 (s, 2H), 3.83 (s, 4H), 3.86 (s, 4H), 6.78 (d, 1H, J = 3.5 Hz), 6.83 (d, 1H, J = 3.6 Hz), 6.91 (d, 2H, J = 3.5 Hz), 6.95 (d, 2H, J = 3.6 Hz), 7.09 (d, 2H, J = 3.5 Hz), 7.37 (d, 2H, J = 3.5 Hz).
13C NMR (125 MHz, (CD3)2SO, δ, ppm): 35.37 (CH2), 35.54 (CH2), 35.62 (CH2), 54.07 (Csp3 fullerene cage), 56.28 (Csp3 fullerene cage), 59.64 (Csp3 fullerene cage), 75.47 (Csp3 fullerene cage-Cl), 126.87, 127.04, 127.45, 127.47, 127.60, 129.88, 137.90, 138.11, 138.45, 139.14, 140.21, 142.26, 142.54, 142.82, 143.10, 143.37, 143.89, 144.29, 144.30, 144.33, 144.61, 144.66, 144.93, 145.60, 146.00, 147.12, 147.26, 147.83, 148.18, 148.23, 148.32, 148.62, 148.69, 148.72, 149.95, 150.58, 153.32, 155.70, 171.79 (COOH), 171.92 (COOH), 171.93 (COOH). FTIR (KBr pellet, ν, cm−1): 540 (M), 588 (M), 618 (M), 646 (M), 692 (M), 1042 (M), 1110 (W), 1236 (M), 1274 (M), 1336 (M), 1380 (S), 1442 (M), 1580 (VS), 1640 (S), 1658(M), 3364 (S), 3386 (S), 3396 (S), 3406 (S).
Compound 2-OMe. (90%)1H NMR (500 MHz, CDCl3, δ, ppm): 1.22 (t, 3H, J = 7.1 Hz), 2.10 (q, 2H, J = 7.1 Hz), 3.72 (s, 3H), 3.75 (s, 6H), 3.75 (s, 6H), 3.77 (s, 2H), 3.85 (s, 8H), 6.82 (d, 1H, J = 3.6 Hz), 6.87– 6.89 (m, 4H), 6.91 (d, 1H, J = 3.6 Hz), 7.11 (d, 2H, J = 3.3 Hz), 7.22 (d, 2H, J = 3.6 Hz).
13C NMR (125 MHz, CDCl3, δ, ppm): 9.62 (CH2CH3), 32.55 (CH2CH3), 35.27 (CH2), 35.53 (CH2), 35.58 (CH2), 52.30 (CH3), 52.31 (CH3), 54.24 (Csp3 fullerene cage), 56.51 (Csp3 fullerene cage), 59.84 (Csp3 fullerene cage), 65.25 (Csp3 fullerene cage), 126.12, 126.61, 126.78, 126.88, 126.90, 130.33, 135.30, 135.42, 135.97, 141.96, 142.57, 142.89, 143.09, 143.36, 143.45, 143.58, 144.17, 144.20, 144.49, 144.53, 144.78, 145.02, 146.03, 146.72, 146.99, 147.19, 147.29, 147.84, 148.12, 148.17, 148.20, 148.56, 148.58, 148.76, 150.69, 152.73, 155.50, 155.76, 170.61 (COOCH3), 170.65 (COOCH3), 170.69 (COOCH3). FTIR (KBr pellet, ν, cm−1): 538 (M), 796 (M), 1004 (M), 1038 (M), 1108 (S), 1168 (S), 1220 (S), 1262 (M), 1434 (S), 1710 (S), 1730 (VS), 1738 (VS), 2364 (M), 2854 (M), 2924 (S), 3430 (S).
Compound 2-OH. (96%)1H NMR (500 MHz, (CD3)2SO, δ, ppm): 1.20 (t, 3H, J = 6.9 Hz), 1.97– 2.06 (m, 2H), 3.76 (s, 2H), 3.84 (s, 8H), 6.82– 6.86 (m, 2H), 6.90– 6.96 (m, 4H), 7.07 (d, 2H, J = 3.4 Hz), 7.21 (d, 2H, J = 3.4 Hz).
13C NMR (125 MHz, (CD3)2SO, δ, ppm): 9.66 (CH2CH3), 32.56 (CH2CH3), 35.34 (CH2), 35.57 (CH2), 35.61 (CH2), 54.31 (Csp3 fullerene cage), 56.63 (Csp3 fullerene cage), 59.91 (Csp3 fullerene cage), 65.20 (Csp3 fullerene cage), 126.21, 126.79, 127.16, 127.39, 127.52, 130.36, 137.38, 137.63, 138.35, 140.37, 141.90, 142.80, 142.90, 143.16, 143.32, 143.61, 144.11, 144.20, 144.45, 144.51, 144.78, 144.88, 144.91, 145.04, 145.42, 146.93, 147.06, 147.16, 147.23, 147.83, 148.09, 148.12, 148.51, 148.72, 151.08, 153.12, 155.58, 156.04, 171.82 (COOH), 171.93 (COOH), 171.98 (COOH). FTIR (KBr pellet, ν, cm−1): 618 (S), 758 (M), 812 (M), 1046 (S), 1116 (VS), 1216 (S), 1380 (S), 1448 (S), 1594 (S), 2344 (S), 2362 (S), 2854 (S), 2924 (VS), 3206 (VS), 3268 (VS), 3310 (VS).
Compound 5-OMe. (40%)1H NMR (600 MHz, CDCl3, δ, ppm): 1.67 (m, 10Н), 1.98 (m, 10Н), 2.61 (t, 2Н), 2.66 (t, 4Н), 2.71 (t, 4Н), 3.38 (t, 1Н), 3.42 (t, 2Н), 3.44 (t, 2Н), 3.73 (s, 6Н), 3.75 (s, 12Н), 3.76 (s, 12Н), 5.24 (s, 1Н), 6.98 (d, 2Н), 7.01 (d, 4Н), 7.15 (d, 4Н), 7.32 (d, 2Н), 7.50 (d, 4Н), 7.67 (d, 4Н).
13C NMR (150 MHz, CDCl3, δ, ppm): 28.35 (CH2), 28.39 (CH2), 28.42 (CH2), 29.00 (CH2), 29.06 (CH2), 34.89 (CH2), 34.95 (CH2), 35.02 (CH2), 51.52 (CH), 52.51 (CH3), 52.54 (CH3), 58.67 (Csp3 fullerene cage), 58.73 (Csp3 fullerene cage), 60.77 (Csp3 fullerene cage), 62.91 (Csp3 fullerene cage), 127.78, 128.08, 128.21, 128.63, 128.78, 128.89, 137.43, 137.47, 140.66, 140.93, 141.13, 143.12, 143.39, 143.81, 144.03, 144.11, 144.23, 144.33, 144.53, 145.47, 145.73, 145.98, 146.23, 146.91, 147.10, 147.20, 147.74, 147.94, 148.06, 148.25, 148.38, 148.67, 148.72, 151.78, 152.37, 152.61, 156.20, 169.78 (СOOCH3).
Compound 5-OH. (96%)1H NMR (600 MHz, (CD3)2CO, CS2, δ, ppm): 1.50–1.75 (m, 10Н), 1.75–1.97 (m, 10Н), 2.57 (t, 2Н), 2.63 (t, 4Н), 2.71 (t, 4Н), 3.27–3.40 (m, 5Н), 5.36 (s, 1Н), 6.99 (d, 2Н), 7.04 (d, 4Н), 7.20 (d, 4Н), 7.25 (d, 2Н), 7.49 (d, 4Н), 7.71 (d, 4Н).
13C NMR (150 MHz, (CD3)2CO, CS2, δ, ppm): 28.19 (CH2), 28.28 (CH2), 28.42 (CH2), 29.79 (CH2), 34.96 (CH2), 35.03 (CH2), 35.15 (CH2), 51.16 (CH), 51.18 (CH), 51.21 (CH), 58.72 (Csp3 fullerene cage), 58.82 (Csp3 fullerene cage), 60.86 (Csp3 fullerene cage), 63.14 (Csp3 fullerene cage), 127.76, 128.13, 128.26, 128.85, 128.95, 129.03, 129.16, 136.80, 136.83, 141.18, 141.40, 141.58, 143.21, 144.04, 144.11, 144.18. 144.20, 144.29, 144.42, 144.78, 144.83, 145.69, 145.98, 146.19, 146.73, 146.95, 147.15, 147.25, 147.76, 148.10, 148.28, 148.31, 148.41, 148.69, 148.73, 152.08, 152.63, 152.77, 156.58, 170.49 (СOOH), 170.58 (СOOH), 170.70 (СOOH).
Compound 6-OMe. (5%)1H NMR (500 MHz, CDCl3, δ, ppm): 3.26 (s, 8H), 3.43 (s, 8H), 3.55 (s, 4H), 3.65 (s, 4H), 3.69 (s, 8H), 3.72 (s, 8H), 6.80 (dd, 2H, J = 8.2, 2.0 Hz), 6.98 (d, 2H, J = 1.9 Hz), 7.15 (dd, 2H, J = 8.1, 2.0 Hz), 7.17 (d, 2H, J = 1.8 Hz), 7.56 (d, 2H, J = 8.2 Hz), 7.98 (d, 2H, J = 8.0 Hz).
13C NMR (125 MHz, CDCl3, δ, ppm): 40.46 (CH2), 40.61 (CH2), 51.90, 52.13, 52.24, 59.75 (Csp3 fullerene cage), 60.15 (Csp3 fullerene cage), 128.22, 128.77, 128.96, 129.15, 131.04, 131.24, 131.50, 133.07, 133.15, 133.95, 134.09, 134.76, 134.80, 136.07, 136.43, 137.40, 140.68, 142.19, 142.32, 142.77, 143.04, 143.46, 143.70, 143.93, 145.06, 145.23, 145.61, 145.68, 146.52, 146.73, 146.85, 147.35, 147.40, 147.50, 148.29, 148.41, 148.82, 149.19, 149.61, 149.78, 152.40, 162.24, 171.60 (COOCH3), 171.67 (COOCH3), 171.77 (COOCH3), 171.91 (COOCH3).
ESI MS: m/z= 1606 ([M]−).
Compound 6-OH. (95%)1H NMR (600 MHz, (CD3)2SO, δ, ppm): 3.47 (s, 8H), 3.60 (s, 8H), 6.75 (d, 2H, J = 8.3 Hz), 6.99 (m, 2H), 7.17– 7.25 (m, 4H), 7.52 (d, 2H, J = 8.2 Hz), 8.04 (d, 2H, J = 8.0 Hz), 12.32 (s, 8H).
13C NMR (125 MHz, (CD3)2SO, δ, ppm): 40.62 (CH2), 40.65 (CH2), 59.42 (Csp3 fullerene cage), 59.79 (Csp3 fullerene cage), 128.35, 129.00, 130.40, 132.31, 133.42, 133.75, 134.55, 134.80, 134.94, 135.43, 136.60, 137.09, 137.32, 139.74, 141.82, 141.89, 142.21, 142.61, 142.81, 143.12, 143.67, 144.72, 144.99, 145.16, 145.44, 145.49, 145.81, 145.85, 146.19, 146.60, 146.81, 147.37, 147.48, 147.78, 147.99, 148.28, 148.47, 148.78, 149.19, 149.91, 152.33, 162.52, 172.36 (COOH), 172.44 (COOH), 172.46 (COOH), 172.52 (COOH).
Compound 7-OMe. (10%)1H NMR (500 MHz, CDCl3, δ, ppm): 3.56 (s, 5H), 3.58– 3.61 (m, 15H), 3.63 (s, 5H), 3.67– 3.71 (m, 25H), 7.08 (m, 2H), 7.18 (d, 1H, J = 7.9 Hz), 7.21 (d, 1H, J = 7.7 Hz), 7.23 (d, 1H, J = 8.6 Hz), 7.25– 7.27 (m, 1H), 7.30 (d, 1H, J = 8.0 Hz), 7.45 (d, 1H, J = 2.1 Hz), 7.49 (d, 1H, J = 2.0 Hz), 7.51 (dd, 1H, J = 7.9, J = 2.1 Hz), 7.54 (dd, 1H, J = 7.8, J = 2.0 Hz), 7.72– 7.74 (m, 3H), 7.86 (dd, 1H, J = 7.9, J = 2.0 Hz).
13C NMR (125 MHz, CDCl3, δ, ppm): 38.36 (CH2), 38.50 (CH2), 38.76 (CH2), 38.78 (CH2), 38.87 (CH2), 52.01 (CH3), 52.08 (CH3), 52.12 (CH3), 52.15 (CH3), 52.22 (CH3), 57.84 (Csp3 fullerene cage), 57.97 (Csp3 fullerene cage), 60.39 (Csp3 fullerene cage), 60.53 (Csp3 fullerene cage), 63.13 (Csp3 fullerene cage), 76.08 (Csp3 fullerene cage-Cl), 127.64, 127.68, 127.85, 128.07, 128.47, 128.65, 130.46, 131.18, 131.24, 131.40, 131.60, 132.61, 132.89, 133.06, 133.17, 133.24, 133.30, 134.01, 134.18, 134.20, 134.22, 134.30, 136.98, 137.27, 138.47, 142.82, 142.85, 142.97, 142.99, 143.18, 143.35, 143.36, 143.82, 143.85, 143.86, 143.90, 144.22, 144.30, 144.40, 144.43, 144.50, 144.72, 144.56, 145.10, 145.26, 145.44, 145.51, 146.57, 146.63, 147.36, 147.39, 147.50, 147.53, 147.94, 148.03, 148.32, 148.60, 148.62, 148.72, 148.82, 148.86, 148.90, 148.93, 148.97, 150.08, 150.67, 150.98, 151.29, 153.44, 154.29, 156.16, 156.62, 171.22 (COOCH3), 171.25 (COOCH3), 171.27 (COOCH3), 171.28 (COOCH3), 171.32 (COOCH3), 171.35 (COOCH3), 171.41 (COOCH3), 171.47 (COOCH3), 171.50 (COOCH3).
ESI MS: m/z= 1827 ([M-Cl]−).
Compound 7-OH. (96%)1H NMR (500 MHz, D2O, δ, ppm): 3.24– 3.32 (m, 4H), 3.35 (s, 4H), 3.28– 3.45 (m, 4H), 3.45– 3.51 (m, 8H), 6.61– 6.67 (m, 2H), 6.77 (dd, 1H, J = 8.0, 1.8 Hz), 6.95–7.05 (m, 2H), 7.08– 7.24 (m, 6H), 7.44 (s, 2H), 7.68 (s, 2H) (for potassium salt form).
13C NMR (125 MHz, (CD3)2SO, δ, ppm): 38.44 (CH2), 38.53 (CH2), 38.57 (CH2), 38.86 (CH2), 38.88 (CH2), 57.79 (Csp3 fullerene cage), 57.85 (Csp3 fullerene cage), 60.46 (Csp3 fullerene cage), 60.60 (Csp3 fullerene cage), 63.12 (Csp3 fullerene cage), 76.24 (Csp3 fullerene cage-Cl), 126.62, 127.34, 127.49, 128.77, 130.51, 130.79, 131.05, 131.16, 131.72, 131.89, 132.03, 132.43, 132.49, 134.26, 134.36, 134.39, 134.52, 134.58, 134.85, 135.29, 135.41, 135.65, 135.69, 135.73, 135.78, 135.81, 137.86, 137.98, 138.80, 142.68, 143.02, 143.07, 143.62, 143.85, 144.14, 144.24, 145.49, 145.56, 145.89, 146.03, 147.13, 147.21, 147.23, 147.27, 147.36, 147.46, 147.73, 147.80, 147.84, 147.93, 148.05, 148.36, 148.43, 148.51, 148.61, 148.70, 151.69, 152.34, 156.21, 159.58, 172.14 (COOH), 172.38 (COOH), 172.45 (COOH), 172.52 (COOH), 172.54 (COOH), 172.77 (COOH), 172.96 (COOH) (for acid form).
Compound 9-OMe. (25%)1H NMR (500 MHz, CDCl3, δ, ppm): 3.67 (s, 2H), 3.70 (s, 4H), 3.73 (s, 4H), 3.74 (s, 3H), 3.76 (s, 6H), 3.78 (s, 6H), 5.21 (s, 1H), 7.25 (d, 2H, J = 8.7 Hz), 7.29 (d, 4H, J = 8.6 Hz), 7.32 (d, 2H, J = 8.6 Hz), 7.42 (d, 4H, J = 8.5 Hz), 7.53 (d, 4H, J = 8.5 Hz), 7.71 (d, 4H, J = 8.5 Hz).
13C NMR (125 MHz, CDCl3, δ, ppm): 36.08 (CH2), 36.29 (CH2), 36.34 (CH2), 52.65 (CH3), 52.68 (CH3), 52.72 (CH3), 58.39 (Csp3 fullerene cage), 58.52 (Csp3 fullerene cage), 60.51 (Csp3 fullerene cage), 62.89 (Csp3 fullerene cage), 127.73, 128.25, 128.59, 128.71, 129.98, 130.08, 130.34, 130.56, 132.60, 134.58, 134.76, 135.04, 138.25, 138.28, 143.28, 143.53, 144.02, 144.17, 144.28, 144.36, 144.45, 145.20, 145.51, 145.70, 146.91, 147.11, 147.20, 147.38, 147.79, 148.15, 148.19, 148.31, 148.44, 148.72, 148.82, 148.86, 151.13, 151.86, 152.42, 155.73, 169.83 (COOCH3), 169.85 (COOCH3).
ESI MS: m/z= 1626 ([M-H]−).
Compound 9-OH. (95%)1H NMR (500 MHz, (CD3)2SO, δ, ppm): 3.77 (s, 2H), 3.79 (s, 4H), 3.85 (s, 4H), 5.77 (s, 1H), 7.12 (d, 2H, J = 8.3 Hz), 7.17 (d, 2H, J = 8.5 Hz), 7.22 (d, 4H, J = 8.4 Hz), 7.36 (d, 4H, J = 8.3 Hz), 7.64 (d, 4H, J = 8.3 Hz), 7.85 (d, 4H, J = 8.3 Hz).
13C NMR (125 MHz, (CD3)2SO, δ, ppm): 35.18 (CH2), 35.48 (CH2), 35.68 (CH2), 58.19 (Csp3 fullerene cage), 58.48 (Csp3 fullerene cage), 60.54 (Csp3 fullerene cage), 62.62 (Csp3 fullerene cage), 128.16, 128.36, 128.44, 128.69, 128.78, 128.84, 128.98, 129.39, 135.80, 136.10, 136.28, 136.44, 136.54, 142.49, 143.05, 143.57, 143.90, 143.97, 143.99, 144.13, 144.21, 144.74, 145.63, 145.85, 145.90, 146.58, 146.74, 146.95, 147.06, 147.56, 147.89, 147.93, 148.08, 148.18, 148.22, 148.50, 148.58, 151.98, 152.68, 153.36, 156.51, 170.82 (COOH), 170.88 (COOH), 170.93 (COOH).
ESI MS: m/z= 1556 ([M-H]−).
Compound 10-OMe. (8%)1H NMR (500 MHz, CDCl3, δ, ppm): 3.74 (s, 3H), 3.78 (s, 3H), 3.81 (s, 6H), 3.84 (s, 6H), 4.46 (s, 2H), 4.48 (s, 2H), 4.65 (s, 4H), 4.70 (s, 4H), 6.01 (d, 2H, J = 8.8 Hz), 6.44 (d, 2H, J = 8.9 Hz), 6.57 (d, 2H, J = 8.8 Hz), 6.79 (d, 4H, J = 8.8 Hz), 6.88 (d, 4H, J = 8.8 Hz), 7.12 (d, 2H, J = 8.8 Hz), 7.51 (d, 4H, J = 8.7 Hz), 7.65 (d, 4H, J = 8.7 Hz).
13C NMR (125 MHz, CDCl3, δ, ppm): 52.14 (CH3), 52.19 (CH3), 52.33 (CH3), 57.26 (Csp3 fullerene cage), 60.94 (Csp3 fullerene cage), 63.29 (Csp3 fullerene cage), 65.01 (CH2), 65.04 (CH2), 65.30 (CH2), 65.34 (CH2), 72.30 (Csp3 fullerene cage), 112.20, 113.56, 114.86, 115.36, 127.04, 127.73, 129.10, 129.61, 129.99, 130.09, 130.21, 130.56, 131.23, 131.69, 132.59, 133.09, 133.38, 135.70, 138.42, 142.41, 142.80, 143.00, 143.85, 144.01, 144.05, 144.26, 144.37, 144.60, 145.33, 145.69, 147.17, 147.30, 147.53, 147.92, 148.24, 148.34, 148.62, 148.66, 148.83, 148.86, 149.85, 154.15, 155.49, 156.02, 157.33, 157.40, 157.67, 158.94, 169.09 (COOCH3), 169.13 (COOCH3), 169.18 (COOCH3).
ESI MS: m/z= 1711 ([M]−).
Compound 10-OH. (98%)1H NMR (500 MHz, (CD3)2CO, δ, ppm): 4.41 (s, 4H), 4.61 (s, 4H), 4.67 (s, 4H), 6.05 (d, 2H, J = 8.2 Hz), 6.46 (d, 2H, J = 7.8 Hz), 6.65 (d, 2H, J = 7.7 Hz), 6.84 (d, 4H, J = 7.9 Hz), 6.94 (d, 4H, J = 7.7 Hz), 7.22 (d, 2H, J = 7.3 Hz), 7.56 (d, 4H, J = 7.7 Hz), 7.71 (d, 4H, J = 7.8 Hz).
13C NMR (125 MHz, (CD3)2CO, δ, ppm): 57.29 (Csp3 fullerene cage), 61.06 (Csp3 fullerene cage), 63.34 (Csp3 fullerene cage), 64.50 (CH2), 64.58 (CH2), 64.62 (CH2), 72.45 (Csp3 fullerene cage), 112.12, 113.74, 115.01, 129.10, 129.42, 129.86, 130.15, 131.28, 131.64, 132.49, 132.71, 135.40, 137.91, 142.73, 142.91, 143.11, 143.72, 143.93, 143.98, 144.25, 144.43, 144.56, 145.39, 145.57, 146.91, 147.22, 147.54, 147.78, 148.12, 148.18, 148.28, 148.54, 148.77, 148.87, 150.15, 154.15, 154.44, 155.75, 156.31, 157.65, 157.73, 157.79, 159.33, 170.03 (COOH), 170.12 (COOH), 170.40 (COOH).
ESI MS: m/z= 1626 ([M-H]−), 812 ([M-2H]2-).
Cell Culture
A NSCLC cell line, A549 (ATCC® CCL-185™), was obtained from American Type Culture Collection (ATCC).27,28 Cells were cultured in the Roswell Park Memorial Institute medium (RPMI 1640, Gibco) with 10% fetal bovine serum (FBS, Gibco), and in a humidified 5% CO2 incubator at 37°C. The subculture process was performed two times every week.
Cell Viability Analysis
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay29 was used to estimate the cytotoxicity of fullerene derivatives. The powder form of all water-soluble fullerene derivatives was freeze-dried for 24 h before cell viability analysis (FDU-1200, EYELA, JPN). Fullerene derivatives were then dissolved in cell culture medium. A549 cells line were seeded into 96-well culture plates in a density of 1500 cells/well and then incubated for 24 h before the exposure to fullerene derivatives. After the incubation, the medium was replaced by cell culture medium containing fullerene derivatives and then incubated in the humidified incubator for 72 h. To perform the MTT assay, 100 μL of MTT in deionized water (0.5 mg/mL, Sigma) was added to each well and then incubated in the incubator for 1 h to generate products of purple formazan. Following 1 h of reaction, 100 μL of dimethyl sulfoxide (DMSO) was added to each well to dissolve the purple formazan products into a colored solution. The colored solution was measured by a multimode microplate reader (SpectraMax® iD3) with the test wavelength at 570 nm. To obtain more accurate cytotoxicity values, CCK-8 assay was carried out to evaluate underestimate compounds. After the incubation of fullerene derivatives with A549 cells for 72 h, 100 μL of CCK-8 regent was added to each well to generate orange formazan dye and detected by a test wavelength at 450 nm. The raw data were analyzed by the tool SoftMax® Pro 7 Software. The IC50 values characterizing cytostatic activity of compounds were calculated by the GraphPad Prism 6 software (Version 6.01). The percentage of cell viability was calculated according to the following equation:
(1)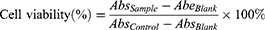
where AbsSample means the optical density of cells with tested compounds, AbsControl indicates the optical density of control cells, and AbsBlank is the absorbance of PBS.
Construction of QSAR Model
A set of 10 fullerene derivatives with measured IC50 values were utilized as a training set or test set for the QSAR model generation. The 3D geometry structure of each fullerene derivative was drawn and constructed by BIOVIA Discovery Studio (BIOVIA, USA). After the optimization of 3D geometry of the structures, a series of descriptors was calculated by the protocol of the Discovery Studio software. The IC50 value of each compound was defined as the dependent property, and the calculated parameters were treated as independent variables in model building. Before the process of QSAR model generation, the measured bioactivity values (IC50) of all fullerene derivatives were converted to the pIC50 values as follows:
(2)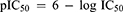
The genetic function approximation (GFA) algorithm30 was used to generate equations of the QSAR model, which contains molecular descriptors correlating with the activity values.
Determination of Reduced Glutathione (GSH)
The health condition of cells after the treatment with fullerene derivatives was examined by using the VitaBright-48™ (VB-48, Chemometec) and propidium iodide (PI, Chemometec) staining. The reagent of VB-48 used to stain healthy cells generates fluorescence after reaction with thiols, that can be detected by the image analysis with Nucleo-Counter NC-3000 (ChemoMetec). To perform the test, A549 cells after exposure to fullerene derivatives for 72 h were washed with PBS. After the steps of trypsinization and centrifugation, cells were stained with VB-48™ and PI for 5 min before the intensities were detected and quantified by Nucleo-Counter NC-3000. Image analysis was processed by the NC-3000 software.
Statistical Analyses
The experimental data measurements were performed in three independent series for each group. All data were displayed as the mean ± standard deviation. The R-squared (q2) of cross-validation to estimate the accuracy and precision of QSAR models was obtained from the following equation:
(3)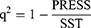
where PRESS indicates the predictive sum of squares of a model, and the SST is the total sum of squares.
Results
Cytotoxicity for Various Water-Soluble Fullerene Derivatives
The chemical structures and biological activities of the water-soluble fullerene derivatives are displayed in Figure 2. The measured IC50 values and standard deviation (SD) were first determined from the results of three independent experiments measured by MTT assay (Figure S38). In MTT assay, compounds 2 and 10 had a significant cytotoxic effect on A549 cells. The IC50 values were 89.16 μM and 75.77 μM, respectively (Table 1). Compounds 1, 3, 4, 8, and 9 also inhibited cell proliferation, and the cell viability decreased in a dose-dependent manner. Meanwhile, compounds 5, 6, and 7 were less cytotoxic with the IC50 values being beyond 400 μM (the highest concentration tested). For the latter three compounds, the IC50 values were estimated by extrapolation of the compound dose-cell viability dependences revealed by MTT assay. Unfortunately, the IC50 values estimated by MTT assay demonstrated large variation in several experiments for compounds 1, 4, 5, 6 and 9.
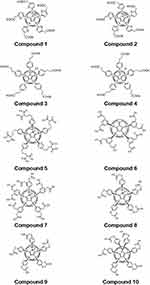 |
Figure 2 The chemical structures of the fullerene derivatives. |
In order to determine IC50 values more precisely, we further utilized the cell counting kit-8 (CCK-8) assay as the second method to measure the cytotoxicity fullerene derivatives on A549 cells. The IC50 values measured by CCK8 assay are listed in Table 1, and the dose–response relationships are shown in Figure S39. As the results indicated, the IC50 values obtained by CCK8 assay were notably greater than those revealed by MTT assay for all tested compounds. Results by CCK8 were not consistent with those of MTT expect for compounds 2, 3, and 6.
Validation of Cytotoxicity Value by Direct Staining
A direct stain by VB-48 and PI was employed to determine which assay gives better IC50 value for these compounds. Results from the VB-48/PI staining are summarized in Figure 3. The data were more consistent with the IC50 values obtained by CCK8 method rather than using MTT assay. According to the results delivered by the CCK8 method, the compound 4 should induce a stronger cytotoxic effect than compounds 2 and 3. Therefore, MTT assay appeared to underestimate the cytotoxic effect of compound 4.
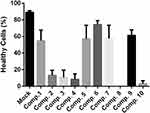 |
Figure 3 Evaluation of cell viability 72 h after their exposure to compounds 1–10. |
QSAR Model Generation
The cytotoxicity data represented by IC50 values obtained using the CCK8 assay were used to build QSAR models for the fullerene derivatives. The chemical structures and the measured cytotoxicity values used for the model generation are listed in Table 1. There were 204 molecular descriptors generated from the QSAR analyzing tools of BIOVIA Discovery Studio. Molecular descriptors of fullerene derivatives related to pIC50 values were identified by GFA algorithm. The top ten equations of QSAR model that correlated with the biological activity are shown in Table 2. The least squares fitting r2 and cross-validated correlation coefficient q2 values were criteria of validating of different models. The QSAR model, which had r2 of 0.9966 and q2 of 0.9246, can be recommended since it gives the best match between the experimental and the predicted cytotoxicity values as shown in Figure 4. The equation of the selected QSAR model is described as follows:
(4)
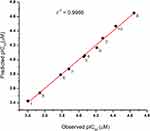 |
Figure 4 Correlation of the observed cytotoxicity in CCK8 assays (pIC50) with the predicted cytotoxicity (pIC50) using the recommended QSAR. |
The descriptors selected by GFA algorithm were S_Count, ES_Count_dO, ES_Count_sCl, HBA_Count, Num_AliphaticSingleBonds, and Num_H_Acceptors and Num_RingBonds. Here, S_Count means the number of sulfur atoms in the molecule of a fullerene derivative. The next two descriptors are relevant to the properties of electrotopological state (E-state values), which include ES_Count_dO, and ES_Count_sCl properties. ES_Count_dO means the oxygen atom with double bond, the carbonyl C=O group. ES_Count_sCl indicates the chlorine atom with single bond. HBA_Count is the number of hydrogen bond accepting groups in a fullerene compound. Finally, Num_AliphaticSingleBonds defines the number of single bonds in saturated hydrocarbons, while Num_H_Acceptors and Num_RingBonds indicates the number of heteroatoms among the hydrogen bond acceptors and ring bonds within entire molecules, respectively. All calculated data of these six descriptors for the ten fullerene derivatives are listed in Table 3, which were ranked by pIC50 values. Compounds 5–7 had low anticancer activities, and these three compounds contained more single bonds than the other candidates in saturated hydrocarbon chains. The values of Num_AliphaticSingleBonds were 91, 73, and 91, respectively. The result indicated that the solubilizing addends such as potassium 2-(3-phenylpropyl)malonate, 2-(3-phenylpropyl) malonic acid, and potassium 2,2ʹ-(1,2-phenylene) diacetate may not be suitable to design efficient inhibitors for A549 lung cancer cells. Compound 10, the most cytotoxic one, has a number 53 in the Num_AliphaticSingleBonds descriptor. Because the compound 10 bears six residues of potassium 2-phenoxyacetate, it seems that a greater number of aromatic bonds have positive effect by increasing the cytotoxicity of fullerene derivatives with respect to A549 cells.
 |
Table 3 The Computed Values of Chemical Properties for Water-Soluble Fullerene Derivatives Using the Recommended QSAR Model Compared to Bio-Activities Provided by CCK8 Assay |
Discussion
Fullerene derivatives are attractive molecules for various biological and medical applications. For instance, water-soluble fullerene derivatives have been studied for the anti-HIV activity as a possible therapy of AIDS.31–33 In other cases, the fullerenes are used as potential radical scavengers to reduce the concentration of reactive oxygen species (ROS).34–36 Nevertheless, there are still few studies on investigation of quantitative structure–activity relationship for fullerene derivatives. The current study examined the relationship between the antitumor effect and the chemical nature of functional groups incorporated in the structures of water-soluble fullerenes using the approach of molecular simulations.
Previously we have investigated four classes of water-soluble fullerene compounds that have different chemical linkages between the fullerene cage and the functional groups: C−C bonds, C−N bonds, C−P bonds, and C−S bonds and compared their antitumor efficacies.19 Our results showed that the fullerene derivatives with the addends bearing a carboxylic group at aromatic ring attached to the cage had no cytotoxicity with respect glioblastoma cells but induced neural stem cell proliferation. Meanwhile, another research group has recently demonstrated that 4-phenylbutanoate enhanced the effects of apoptosis and endoplasmic reticulum stress in human lung cancer cells induced by cisplatin,37 and the cell cycle of human gastric cancer cells can be arrested at the G0/G1 phase after treatment with 4-phenylbutanoate.38 These findings suggested that fullerene derivatives with 4-phenylbutanoate groups attached to the carbon cage might have antitumor effects on human cancer cells. The phenylbutanoate has one more carbon atom than phenylpropanoate in the aliphatic chain. To the best of our knowledge, there are no reports on anticancer effects of compounds bearing residues of phenylpropanoate. For the synthesis of the water-soluble fullerene derivatives, we considered the phenylpropanoate as one possible functional group to be attached to the fullerene cage and further investigated for the cytotoxicity on human lung cancer cells.
The MTT assay was first employed in the current study to determine the IC50 value of each functionalized fullerene derivative on A549 cancer cells. Both compounds 3 and 4 incorporate five functional groups of 3-phenylpropanoate at R2 and R3 positions (Table 1, Figure 2). We found that compound 3 had a cytotoxic effect on A549 cells with an IC50 value of 133.73 μM. However, compound 4 revealed much lower cytotoxicity with the IC50 value of 203.97 μM. We further observed the structure–activity relationship for compounds 1–4 based on the results of MTT test. The chlorine atom at R1 position reduced the inhibitory effect for compound 1 as compared to Et in compound 2. However, introduction of hydrogen at R1 position in case of compound 4 leads to further increase of IC50 value (weakening of anticancer action) as compared to 3 bearing Cl. The inconsistent MTT results as well as large standard errors of IC50 values suggested that the MTT assay may not be a proper tool to estimate IC50 for certain fullerene derivatives.
We utilized two alternative methods, the CCK-8 assay and VB-48/PI staining to obtain more accurate IC50 values for the fullerene derivatives. The inhibitory effect determined by CCK-8 assay was more consistent with the results of live/dead cell VB-48/PI staining. Therefore, CCK-8 assay should be considered as a more appropriate approach to determine IC50 values for the fullerene derivatives rather than the MTT assay.
We further analyzed the key features of the fullerene derivatives responsible for their cytotoxic effect. Note that compounds 1 and 3 each have chlorine atom at R1 position, while compounds 1 and 2 have the same functional groups at R2 and R3 positions. Compound 4 has a hydrogen atom at R1 position, while the chemical structures of compounds 3 and 4 are similar due to the same attached groups at R2 and R3 positions. For the structure–activity relationship of compounds 1–4, the chlorine atom at R1 position increased significantly the IC50 values of the fullerene derivatives for A549 cell line. On the contrary, hydrogen atom or ethyl group attached at R1 position enables enhanced cytotoxicity of the fullerene derivatives.
Interestingly, some fullerene derivatives explored in the current study could increase cell proliferation when applied in concentrations between 25 μM and 50 μM, as revealed both by both MTT and CCK-8 assays. These findings seem to be consistent with the previous study of Dugan et al,39 who demonstrated that fullerene has antioxidant ability and eliminates efficient superoxide radicals, which enhances cell viability. In addition, soluble fullerene derivatives have potentially short-time antioxidant effects.40 Our data suggested that some of the fullerene derivatives developed in this work might also demonstrate free radical scavenging activity and improve cell viability at low concentrations.
The QSAR approach developed in this work can be further extended to rationalize antitumor activity of the fullerene derivatives reported previously. Moreover, we have employed the experimental data of fullerene compounds from the previous report to confirm the QSAR models are reasonable in this study. Two different types of fullerene derivatives were found to be cytotoxic to A549: compounds with C−N (F1-F7) and C−S (F8-F10) linkages between the solubilizing functional addends and the fullerene cage.41 However, IC50 values were not determined in the previous work. As a matter of fact, even in the current study the IC50 values of the ten compounds used for building QSAR model were obtained from testing a total of 22 fullerene derivatives, of which 12 compounds did not show IC50 values after evaluation. In the previous work, the cell viability was measured at the concentration of 200 μM for all different fullerene derivatives. Using a modified approach, we utilized the cell viability data instead of IC50 values from the previously published work of fullerene derivatives (F1-F10) to generate an alternative QSAR model (Supplementary results) and identify solubilizing functional groups that enable the observed biological activities of these compounds (Table S1). By this effort, we could test the reliability of the suggested descriptors obtained from the recommended equation model on a different series of fullerene derivatives (Figure S40). Notably, the most reasonable QSAR model equation generated to describe previously reported activities of compounds F1-F10 contained the same properties as the QSAR model developed for compounds 1–10 in this work (Table S2). For instance, such descriptors as ES_Count_sCl and Num_AliphaticSingleBonds appear to be important for both series of compounds. Taken together, the findings indicate that the chlorine atom and the number of aliphatic single bonds in the functional groups of the water-soluble fullerene derivatives somehow directly correlate with the activity of these compounds against A549 lung cancer cells. In particular, lower number of aliphatic single bonds corresponds to the greater antitumor activities. Thus, compounds 2, 8, 9, and 10 with just a single CH2 group linking COOK to the aromatic ring (directly or via additional O or S atoms) have stronger anticancer effects than the other fullerene derivatives with longer aliphatic spacers (eg compound 5). Compounds 2, 9, and 10 have no chlorine atoms and reduced number of aliphatic single bonds compared to the others. Besides, the result shown that the greater number of aromatic rings enhanced the cell viability of A549. Compounds 8 and 9 have similar structures but very different cytotoxic concentrations. Compound 9 has sulfur linkage (-SCH2-), which might be responsible for the lower antitumor activity. The most cytotoxic fullerene derivative 8 has OCH2 linkage, which might be responsible for its antitumor efficacy.
It should be emphasized that, to the best of our knowledge, we report the first attempt to correlate the experimentally revealed anticancer effects of water-soluble fullerene derivatives to the peculiarities of their molecular structure through QSAR analysis. QSAR approach was previously used to model and understand the anti-HIV effects of the fullerene derivatives.42 However, these studies should not be focused on a single specific viral protein such as HIV protease, which is believed to be the target for the fullerene-based anti-HIV therapy.32 For instance, Castro et al challenged the validity of the protein active center docking methods by proving that the actual HIV inhibition mechanism is not related to the HIV protease inactivation.42 QSAR is not a target-dependent approach, though it has certain limitations particularly with respect to understanding the role and action mechanisms of the screened bioactive compounds. The advantage of QSAR model is the easiness to find out potential properties from a series of chemical derivatives. However, the disadvantage of QSAR approach has difficulty on establishing a reasonable model through appropriate properties. Castro et al pointed out C60 and C70 fullerene derivatives performed different mechanisms of anti-HIV inhibition due to the difference in aqueous solubility.43 It is necessary to consider the solubility of fullerene derivatives as an important factor during the process of QSAR model generation for anti-HIV drug design, which may help to overcome the limitations of the QSAR approach. The QSAR model developed in this study reveals very useful relationships between the chemical structure of the compounds and their antitumor effects, which can help to optimize the design of water-soluble fullerene derivatives for more efficient anticancer drug.
Conclusion
We synthesized and investigated a series of water-soluble fullerene derivatives that displayed a range of cytotoxic effects on A549 cancer cells. We established a QSAR model to identify the potential key features in the chemical structure of the fullerene derivatives enabling their anticancer action. The structure–activity relationships identified that the more cytotoxic fullerene derivatives had a lower number of aliphatic single bonds in attached solubilizing addends. Moreover, the 2-phenoxyacetate residues and no chlorine atoms are incorporated in the structures of compounds most effectively inhibiting the cancer cell growth. The current work demonstrates that QSAR modeling can significantly facilitate the understanding of the biological effects induced by the functional solubilizing addends in water-soluble fullerene derivatives, particularly the cytotoxicity of these compounds with respect to lung cancer cells. The obtained results may be used for performing a rational design of the surface functional groups of water-soluble fullerene derivatives for more efficient treatment of lung cancer in the future.
Abbreviations
NSCLC, non-small cell lung cancer; SCLC, small-cell lung carcinoma; QSAR, quantitative structure–activity relationship; CADD, computer-aided drug design; IC50, half-lethal inhibition concentrations; SD, standard deviation; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; VB-48, VitaBright-48™; PI, propidium iodide; GFA, genetic function approximation; ROS, reactive oxygen species.
Acknowledgments
This research was supported by the bilateral Taiwanese-Russian research project and Ministry for Science and Education of the Russian Federation (RFBR No. 16-53-52030; MOST 105-2923-E-002−003-MY3), the research plan of National Taiwan University Core Consortium (NTU-CC-109L891001), and the Program for Additive Manufacturing (MOST 108-2811-E-002-554). We also acknowledge the funding of the National Health Research Institutes for the postdoctoral fellow. OAK and PAT acknowledge a general support from the Ministry of Science and Education of the Russian Federation (project No. 0089-2019-0010 / AAAA-A19-119071190044-3). We are grateful to Dr. Yuh-Shan Jou, Academia Sinica, for his assistance in providing the purchased A549 cells.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi:10.3322/caac.20073
2. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi:10.1016/S0025-6196(11)60735-0
3. Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644. doi:10.1016/j.ccm.2011.09.001
4. Li Y, Li Y, Liu J, et al. Expression levels of microRNA-145 and microRNA-10b are associated with metastasis in non-small cell lung cancer. Cancer Biol Ther. 2016;17(3):272–279. doi:10.1080/15384047.2016.1139242
5. Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016;35(1):75–91. doi:10.1007/s10555-016-9618-0
6. Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. doi:10.1056/NEJMoa011954
7. Padma VV. An overview of targeted cancer therapy. BioMedicine. 2015;5(4):19. doi:10.7603/s40681-015-0019-4
8. Feng D, Leong M, Li T, Chen L, Li T. Surgical outcomes in patients with locally advanced gastric cancer treated with S-1 and oxaliplatin as neoadjuvant chemotherapy. World J Surg Oncol. 2015;13:11. doi:10.1186/s12957-015-0444-6
9. Ahn SH, Hong HJ, Kwon SY, Kwon KH, et al; Korean Society of T-H, Neck Surgery Guideline Task F. Guidelines for the surgical management of laryngeal cancer: Korean Society of Thyroid-head and Neck Surgery. Clin Exp Otorhinolaryngol. 2017;10(1):1–43. doi:10.21053/ceo.2016.01389
10. Tonge PJ. Drug-target kinetics in drug discovery. ACS Chem Neurosci. 2018;9(1):29–39. doi:10.1021/acschemneuro.7b00185
11. Tang J, Karhinen L, Xu T, et al. Target inhibition networks: predicting selective combinations of druggable targets to block cancer survival pathways. PLoS Comput Biol. 2013;9(9):e1003226. doi:10.1371/journal.pcbi.1003226
12. Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi:10.1186/s12915-014-0094-0
13. Krzyzosiak A, Sigurdardottir A, Luh L, et al. Target-based discovery of an inhibitor of the regulatory phosphatase PPP1R15B. Cell. 2018;174(5):1216–28 e19. doi:10.1016/j.cell.2018.06.030
14. Salata O. Applications of nanoparticles in biology and medicine. J Nanobiotechnol. 2004;2(1):3. doi:10.1186/1477-3155-2-3
15. Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009;9(5):1909–1915. doi:10.1021/nl900031y
16. Kroto HW, Heath JR, O’Brien SC, Curl RF, Smalley RE. C60: buckminsterfullerene. Nature. 1985;318:162. doi:10.1038/318162a0
17. Wu G, Gao XJ, Jang J, Gao X. Fullerenes and their derivatives as inhibitors of tumor necrosis factor-alpha with highly promoted affinities. J Mol Model. 2016;22(7):161. doi:10.1007/s00894-016-3019-8
18. Bosi S, Da Ros T, Spalluto G, Prato M. Fullerene derivatives: an attractive tool for biological applications. Eur J Med Chem. 2003;38(11–12):913–923. doi:10.1016/j.ejmech.2003.09.005
19. Hsieh FY, Zhilenkov AV, Voronov II, et al. Water-soluble fullerene derivatives as brain medicine: surface chemistry determines if they are neuroprotective and antitumor. ACS Appl Mater Interfaces 2017;9(13):11482–11492. doi:10.1021/acsami.7b01077
20. Yu W, MacKerell AD. Computer-aided drug design methods. Methods Mol Biol. 2017;1520:85–106.
21. De Simone A, Russo D, Ruda GF, et al. Design, synthesis, structure-activity relationship studies, and three-dimensional quantitative structure-activity relationship (3D-QSAR) modeling of a series of O-biphenyl carbamates as dual modulators of dopamine D3 receptor and fatty acid amide hydrolase. J Med Chem. 2017;60(6):2287–2304. doi:10.1021/acs.jmedchem.6b01578
22. Aguiar ACC, Panciera M, Simao Dos Santos EF, et al. Discovery of marinoquinolines as potent and fast-acting plasmodium falciparum inhibitors with in vivo activity. J Med Chem. 2018;61(13):5547–5568. doi:10.1021/acs.jmedchem.8b00143
23. Burello E. Review of (Q)SAR models for regulatory assessment of nanomaterials risks. NanoImpact. 2017;8:48–58. doi:10.1016/j.impact.2017.07.002
24. Troshina OA, Troshin PA, Peregudov AS, Kozlovskiy VI, Balzarini J, Lyubovskaya RN. Chlorofullerene C60Cl6: a precursor for straightforward preparation of highly water-soluble polycarboxylic fullerene derivatives active against HIV. Org Biomol Chem. 2007;5(17):2783–2791. doi:10.1039/b705331b
25. Kraevaya OA, Peregudov AS, Troyanov SI, et al. Diversion of the Arbuzov reaction: alkylation of C–Cl instead of phosphonic ester formation on the fullerene cage. Org Biomol Chem. 2019;17:7155-7160. doi: 10.1039/C9OB00593E
26. Fedorova NE, Klimova RR, Tulenev YA, et al. Carboxylic fullerene C60 derivatives: efficient microbicides against herpes simplex virus and cytomegalovirus infections in vitro. Mendeleev Commun. 2012;22(5):254–256. doi:10.1016/j.mencom.2012.09.009
27. Huang YJ, Hsu SH. Acquisition of epithelial-mesenchymal transition and cancer stem-like phenotypes within chitosan-hyaluronan membrane-derived 3D tumor spheroids. Biomaterials. 2014;35(38):10070–10079. doi:10.1016/j.biomaterials.2014.09.010
28. Yeh HW, Hsu EC, Lee SS, et al. PSPC1 mediates TGF-beta1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol. 2018;20(4):479–491. doi:10.1038/s41556-018-0062-y
29. Mossmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth. 1984;65:55–63. doi:10.1016/0022-1759(83)90303-4
30. Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci. 1994;34(4):854–866.
31. Martinez ZS, Castro E, Seong CS, Ceron MR, Echegoyen L, Llano M. Fullerene derivatives strongly inhibit HIV-1 replication by affecting virus maturation without impairing protease activity. Antimicrob Agents Chemother. 2016;60(10):5731–5741. doi:10.1128/AAC.00341-16
32. Strom TA, Durdagi S, Ersoz SS, Salmas RE, Supuran CT, Barron AR. Fullerene-based inhibitors of HIV-1 protease. J Pept Sci. 2015;21(12):862–870. doi:10.1002/psc.2828
33. Dabrowska A, Pienko T, Taciak P, et al. Fullerene derivatives of nucleoside HIV reverse transcriptase inhibitors-in silico activity prediction. Int J Mol Sci. 2018;19(10):3231. doi:10.3390/ijms19103231
34. Sumi N, Chitra KC. Fullerene C60 nanomaterial induced oxidative imbalance in gonads of the freshwater fish, Anabas testudineus (Bloch, 1792). Aquat Toxicol. 2019;210:196–206. doi:10.1016/j.aquatox.2019.03.003
35. Hazrati MK, Javanshir Z, Bagheri Z. B24N24 fullerene as a carrier for 5-fluorouracil anti-cancer drug delivery: DFT studies. J Mol Graph Model. 2017;77:17–24. doi:10.1016/j.jmgm.2017.08.003
36. Huang YY, Rajda PJ, Szewczyk G, et al. Sodium nitrite potentiates antimicrobial photodynamic inactivation: possible involvement of peroxynitrate. Photochem Photobiol Sci. 2019;18(2):505–515. doi:10.1039/C8PP00452H
37. Shi S, Tan P, Yan B, et al. ER stress and autophagy are involved in the apoptosis induced by cisplatin in human lung cancer cells. Oncol Rep. 2016;35(5):2606–2614. doi:10.3892/or.2016.4680
38. Li LZ, Deng HX, Lou WZ, et al. Growth inhibitory effect of 4-phenyl butyric acid on human gastric cancer cells is associated with cell cycle arrest. World J Gastroenterol 2012;18(1):79–83. doi:10.3748/wjg.v18.i1.79
39. Dugan LL, Turetsky DM, Du C, et al. Carboxyfullerenes as neuroprotective agents. Proc Natl Acad Sci U S A. 1997;94(17):9434–9439. doi:10.1073/pnas.94.17.9434
40. Sergeeva V, Kraevaya O, Ershova E, et al. Antioxidant properties of fullerene derivatives depend on their chemical structure: a study of two fullerene derivatives on HELFs. Oxid Med Cell Longev. 2019;2019:13. doi:10.1155/2019/4398695
41. Wong C-W, Zhilenkov AV, Kraevaya OA, Mischenko DV, Troshin PA, Hsu SH. Toward understanding the antitumor effects of water-soluble fullerene derivatives on lung cancer cells: apoptosis or autophagy pathways? J Med Chem. 2019;62(15):7111–7125. doi:10.1021/acs.jmedchem.9b00652
42. Ibrahim M, Saleh NA, Elshemey WM, Elsayed AA. Fullerene derivative as anti-HIV protease inhibitor: molecular modeling and QSAR approaches. Mini Rev Med Chem. 2012;12(6):447–451. doi:10.2174/138955712800493762
43. Castro E, Martinez ZS, Seong CS, et al. Characterization of new cationic N,N-Dimethyl[70]fulleropyrrolidinium iodide derivatives as potent HIV-1 maturation inhibitors. J Med Chem. 2016;59(24):10963–10973. doi:10.1021/acs.jmedchem.6b00994
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


