Back to Archived Journals » Advances in Genomics and Genetics » Volume 5
From caveman companion to medical innovator: genomic insights into the origin and evolution of domestic dogs
Received 28 February 2015
Accepted for publication 11 April 2015
Published 12 June 2015 Volume 2015:5 Pages 239—255
DOI https://doi.org/10.2147/AGG.S57678
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr John Martignetti

Heidi G Parker, Samuel F Gilbert
National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA
Abstract: The phenotypic and behavioral diversity of the domestic dog has yet to be matched by any other mammalian species. In their current form, which comprises more than 350 populations known as breeds, there is a size range of two orders of magnitude and morphological features reminiscent of not only different species but also different phylogenetic families. The range of both appearance and behavior found in the dog is the product of millennia of human interference, and though humans created the diversity, it remains a point of fascination to both lay and scientific communities. In this review, we summarize the current understanding of the history of dog domestication based on molecular data. We examine the ways that canine genetic and genomic studies have evolved and look at examples of dog genetics in the light of human disease.
Keywords: dog, population, GWAS, mapping, comparative genetics
The origin of the domesticated dog
The origin and history of the domestic dog has been a topic of interest to humans for centuries. As the earliest domesticated animal, our association with the dog far predates our historical records; therefore, the circumstances surrounding our initial meeting and collaboration must be deduced from data that we can obtain through scientific means. The archeological record places the remains of dogs or proto-dogs in sites in Belgium and Siberia more than 30,000 years ago (ya).1,2 Evidence from mitochondrial DNA (mtDNA) sequences suggests that these findings represent domestic lines that died out and do not contribute to our modern dogs; however, analysis of the morphometrics of the skulls suggests that they are extinct wolf lineages rather than dogs.3–5 Fossils that are molecularly or phenotypically more similar to modern dogs have been found in the Middle East and Europe dating 14,000–17,000 ya.3,6 This creates a gap in the archeological record to time the first domestication event. The conditions may have been right for canine domestication as much as 30,000 ya, and it was achieved at least 15,000 ya. Experiments with selection on tameness in foxes have shown that under rigorous selective pressures, phenotypic changes can be accomplished within 20 generations.7 Phenotypic changes may have happened quickly in the dog if we assume that humans had an active role in the domestication process. However, if we assume the domestication of the dogs came from a more gradual move into the human niche, it may have taken much longer to significantly alter their appearance. Because it is possible that the process was repeated multiple times and in multiple locations, the exact temporal placement of the episode or episodes that created our modern dogs is most likely somewhere in between 15,000 ya and 30,000 ya.
Molecular studies have attempted to date the divergence of dog from wolf using multiple data sets and methodologies. mtDNA analyses have timed the divergence of the dog from wolf between 5,400 ya and >100,000 ya.3,8–10 Two recent studies that examined whole-genome sequences of both dogs and wolves estimate the divergence time at 13,000–32,000 ya.11,12 The difficulty in establishing an exact time of domestication stems from multiple sources of conflict. First, the mutation rate of the dog genome is not known. The divergence times are often estimated based on assumed mutation rates, which vary with each study (ie, 1.4×10–8 vs 2.2×10–9 mutations per base pair per generation).11,12 Additionally, wolf–dog introgression must be factored into the calculations. During early domestication, dogs and wolves would have been in close contact and fully capable of interbreeding. Examination of mitochondrial haplotypes and nuclear microsatellite markers in a collection of dogs and wolves from the Caucasus in Georgia indicates that 13% of dogs and 10% of wolves show evidence of hybridization.13 Pervasive hybridization throughout the natural history of the domestic dog confounds attempts at precise estimates of divergence, making dogs and wolves appear more similar and therefore more recently diverged. Finally, the genome of the domestic dog, as a close companion of humans, has been consistently altered through intensive selective pressures to obtain specialized traits.14,15 Thus, while the molecular clock works well in determining species divergence in wild populations, assuming a steady rate of mutation could produce misleading results for a genome in which large regions are being driven to fixation and purifying selection is prevented.16 As we continue to gather more complete genomic information about the full range of dogs and their wild progenitors, we will be able to take all of these factors into account in our search for a precise answer to the time of first domestication.
In the mean time, there are other directions that can be explored to understand early domestication. The large influx of whole-genome sequence data that are currently being produced offers the opportunity to examine the functional genetic alterations that have developed between domesticates and their closest wild relatives. Recent studies have taken this route to identify the genes that were disrupted when dogs became domesticated from wolves.11,17–23 In a comparison of 60 domestic purebred dogs and 12 pooled wolf DNAs, 36 regions of the genome containing 122 genes were found to be under strong selection in the dogs. By mining the gene ontology database, three main gene groups were significantly highlighted: nervous system development, sperm competition, and metabolism/digestion.17 Specifically, under the metabolism heading, three genes involved in starch metabolism stood out. One of these was a copy number increase in dogs (4–30 compared to two in wolves) that increased the expression of the AMY2B (alpha-2B-amylase) gene. The authors hypothesized that the change in diet from largely meat to more plant-based food sources was a primary contributor to domestication, supporting the idea that wolves were first domesticated by scavenging around human agricultural settlements.18 Further investigation of the AMY2B locus, however, shows that the copy number variant (CNV) is not increased in all dog breeds and is polymorphic in some wolves.11 Rather than driving domestication, the AMY2B CNV may be a marker of more recent adaptation to a high-starch diet. A similar, though less dramatic, association has been identified in humans from high starch-consuming populations and the AMY1 genes.19
A second study comprising whole-genome data from three wolves and ten dogs identified fixed clusters of single-nucleotide polymorphisms (SNPs) in each species. Using this method, the study found 204 genes with at least six fixed sequence changes and significantly reduced nucleotide diversity.20 The gene ontology terms that were most significantly overrepresented in this set of genes were behavioral, including response to stress, fear, and defense. These findings were verified in a second set of three wolves and three indigenous dogs. The overrepresentation of behavioral genes in the fixed differences between dogs and wolves suggests that a reduced fear response may have been the driver of early domestication allowing the proto-dog to live comfortably in close contact with early humans.
Finally, an analysis of the sequence of four purebred dogs, four wolves, and four indigenous dogs identified 311 genes under positive selection in the dogs.12 These genes largely represent three gene ontology groups that are identical or nearly identical to those identified in the first study: reproductive, neurological processes, and metabolism/digestion. The authors compared these genes to those under selection in humans and found 32 in common, including genes involved in neurological processes, metabolism, and cancer. The common sites of selection across the genomes of both species indicate similar selective pressures, which is likely the result of our shared environments and constant close interaction. This parallel selection can be seen in the sequence comparison of dogs from high, mid, and low altitudes in the People’s Republic of China.21 The dogs that come from the highest altitudes differed from those of low-altitude regions over the EPAS1 gene and a beta-hemoglobin cluster. EPAS1 is a transcription factor that activates in response to hypoxic conditions or oxygen deprivation and has been identified as a target of selection in the Tibetan highlanders.22,23
Selection on both the AMY2B polymorphism and the EPAS1 gene highlights the convergent selective pressures found in dogs and humans and encourages the further use of dog genetics to inform human evolutionary history. The finding that disease-associated genes, such as those that lead to cancer development, are under selection in both species further strengthens the commonly held hypothesis that disease mutations mapped in dogs will be directly applicable to human-disease studies.24
Techniques used in the study of canine genetics and genomics
The techniques researchers have used to study canine genomics have evolved with the field. Early insights into canine genetics came from pedigree analysis and trait observation. These methods were used to determine the inheritance pattern of physical traits. Physical characteristics of dogs such as blood pressure, pupil diameter, and rectal temperature were attributed to heredity through studies of different breeds of purebred dogs. In a pioneering study, such methods were used to demonstrate that genetic factors, and not just environment, are main contributors to diversity across dog breeds.25
Karyotyping and cytological methods became popular in the 1960s and were used to study a number of aspects about the dog including, but not limited to, sexual abnormalities,26 reproduction,27 and disease.28 Cytogenetics found a resurgence in molecular biology with the advent of more specific probes that allow for chromosome painting and fluorescence in situ hybridization. These methods were used to compare the arrangement of the dog’s small, acrocentric chromosomes to the human genome and showed that gene order is highly conserved between the two.29,30 Further refining the probes has also allowed for more precise identification of chromosome abnormalities and genomic rearrangements.31
Proteins were the focus of canine studies during the 1970s. Many investigations focused on blood diseases or traits such as the clotting disorder von Willebrand disease. Further information on canine biology and its similarities to human was gained through comparison of protein functions across species such as the catalytic properties of enolase,32 retinol transport in plasma,33 and pituitary dosage curves of prolactin,34 which implied a similar structure and function between the dog and human proteins.
New molecular techniques in the 1980s allowed for direct assays of the DNA sequences improving the ability to make comparisons between dogs and humans. Southern blot analysis using cDNA from human leukocyte antigen genes as a probe was able to detect dog leukocyte antigen gene regions, indicating a high level of similarity in the gene sequences of both species’ major histocompatibility complex35 and highlighted the importance of the canine in modeling human diseases. The similarity in dog and canine immune systems was irreplaceable in developing strategies to overcome graft-versus-host disease in bone marrow transplantation and will likely play a future role in disease therapeutics.36–38 It was not until sequencing methods became more automated, however, that large-scale homology studies between dog and human became feasible.
Genetic maps of the dog were created once marker genotyping and analysis methodologies improved in the 1990s.39 Microsatellites, or short tandem repeats, are sequences of DNA that can be identified through restriction enzyme assays or polymerase chain reactions and are the backbone of linkage maps.40–42 The identification of a large number of highly polymorphic microsatellites enabled the construction of the first genetic maps of the dog genome.43,44 These genetic maps made possible the discovery of many genes involved in disease and phenotype determination (reviewed in Parker et al24) and produced the scaffold for assembly of the first complete genome sequence of the dog. The most recent comprehensive linkage map was developed based on the completed sequence specifically to determine recombination rates, given the small size of the dog chromosomes.45
The release of the 7.5× whole-genome sequence of the dog in 2005 represented the culmination of the preceding work on characterizing the dog genome.39 The assembled reference sequence was that of a Boxer and was done through a whole-genome shotgun approach, in which the DNA was fragmented into smaller, clonable segments that allowed for Sanger sequencing. With the assembly of a dog genome sequence, over two million SNPs were identified enabling genome-wide association studies (GWASs). These became a high-powered tool to study dog disease and morphology.
GWASs compare a large number of SNPs across the entire genome of two disparate groups, usually affected cases and unaffected controls for categorical traits, or a range of phenotype values for quantitative traits. Statistical methods are used to identify differences between the groups that associate with the affected status. Such studies have been successful in identifying variants in a number of genes as well as in suggesting candidate regions for specific traits (reviewed in Parker et al24 and Karlsson and Lindblad-Toh46).
SNP genotyping has also been a key in allowing for other genome-wide analyses such as homozygosity mapping and identity-by-descent mapping, in which areas of the genome are compared across multiple individuals to identify common regions of shared alleles.15,47 Both methods can be successfully used to identify genomic regions that are under selection for specific traits or to identify recessive disease traits. For example, homozygosity mapping of six Spinone Italiano with spinocerebellar ataxia and six healthy controls of the same breed identified a disease locus on chromosome 20 that was further narrowed to a repeat expansion in ITPR1.48 This gene has also been associated with ataxias in humans (Table 1).
Haplotype mapping is another powerful tool that employs SNPs to establish a pattern of variants that are commonly inherited. A combination of homozygosity mapping and haplotype comparisons was used to fine map the region associated with chondrodysplasia in a multi-breed GWAS.49 A 24 kb region on canine chromosome 18 (CFA18) was found to be homozygous in 26 dogs from eight chondrodysplastic breeds. All breeds carried an identical haplotype indicating a common ancestral source of the mutation, an fgf4 retrogene inserted in the homozygous region. This technique has been used successfully multiple times in both disease and morphology mapping (Figure 1).
  | Figure 1 Examples of homozygosity and haplotype mapping from studies of morphology and disease phenotypes. |
There are now many canine sequences available due to the advent of next-generation sequencing technologies. The new sequences have been used to identify additional SNPs such as the 4.6 million found by comparing the Korean Jindo sequence to the publically available Boxer and the 1.5× Poodle sequences,50 and to identify insertion/deletions and structural variants.51 The relative low cost and general availability of next-generation sequencing allow for large-scale sequencing to supplement GWASs and linkage studies when searching for disease-associated variants. For instance, a GWAS of patellar luxation in Flat-Coated Retrievers identified four distinct regions of association.52 Instead of fine mapping each of the loci or picking candidate genes, custom genomic hybridization arrays were used to enhance the coding regions in all loci for targeted resequencing in 15 cases and 15 controls. SNPs with the largest difference in frequency between cases and controls were genotyped on a larger panel revealing eight genes that appear associated with the disorder.
Similarly, targeted next-generation sequencing was used to investigate a region on CFA4 that was associated with ataxia in both GWASs and linkage analyses of Old English Sheepdogs.53 The sequencing combined with genotyping and haplotype comparisons in additional affected breeds identified a missense mutation in the RAB24 gene. In contrast, whole-genome sequence of a single Chinese Crested dog with neuronal ceroid lipofuscinosis was compared to the sequence of 101 unaffected dogs of other breeds to identify what was predicted to be a rare disease-causing variant. A deletion was identified in the MFSD8 gene that was homozygous in the diseased dog, absent from the other 101 dogs, and predicted to alter protein structure.54
Next-generation sequencing can enhance the ability to identify variants that are resistant to traditional sequencing or genotyping methods. Targeted next-generation sequencing of a locus from a GWAS in Tibetan Spaniels with late-onset progressive retinal atrophy identified a short interspersed nuclear element (SINE) insertion in an intron of the FAM161A gene that creates a reading frame shift and exon skipping.55 SINE insertions are not annotated in the public sequence databases and can only be identified through exploratory sequencing. Targeted sequencing of the entire associated region proved instrumental in finding this causative mutation.
As it stands today, the dog reference genome is on its third and most updated derivation, CanFam3.1, which includes approximately 2.41 billion base pairs across the 38 autosomal chromosome pairs, the X chromosome, and the mtDNA. The available data have led to the development of highly informative tools for genome analysis in the dog including a high-density SNP chip, expression chips, and a whole-exome enrichment array, which offers a less-expensive alternative to whole-genome sequencing.47,56 The addition of RNAseq data from ten canine tissues has improved the annotation of the canine genome with the current version displaying 20,700 protein-coding genes, 4,600 antisense transcripts, and 7,200 noncoding RNA transcripts positioned confidently and available for mutation analysis.57 Some of the most common resources discussed above can be found in Table 2. The next step in genome annotation will be to identify the regulatory regions, as there is likely to be a subset unique to the dog that will be invaluable for disease studies.
Phenotypic diversity among breeds
By far, the most intriguing aspect of canine genetics is the chance it offers to understand the source of extreme phenotypic diversity (Figure 2). The dog, as we know it today, is the result of centuries of controlled breeding to obtain specialized traits and behaviors. The physical differences between the breeds have long been recognized, but it is through genomic analysis that we have been able to understand the differences at a genetic level. Shortly after the first microsatellites were discovered in the dog, they were being used to assess differences between the breeds. These early studies, often analyzing small numbers of markers and dogs, revealed differences that appeared breed related.58–60 In 2004, a study was released that examined nearly 400 dogs from 85 breeds at 96 microsatellite markers. This study showed definitively that the breeds, with the exception of a single pair, were genetically distinguishable.61 In addition, the study showed that there were four primary clusters of breeds that shared genetic similarity with one another. The first was a group of breeds that were developed in Asia and Africa, the second consisted of mastiff-type breeds, the third cluster combined sighthounds with herding breeds, and the fourth (and the largest) was that of sporting/hunting and companion dogs. These breed clusters grouped on similar physical traits, behavioral traits, or geographic origin. When the study was expanded to over 100 breeds, a fifth breed cluster of large mountain dogs was identified.62
With the development of large-scale SNP genotyping technologies came a large multi-breed study that examined breed relationships and phenotype diversity.63,64 Nearly 50,000 SNPs were genotyped on 912 dogs and 225 wolves collected worldwide. Using a phylogenetic analysis of the SNP genotypes and haplotypes built from the SNPs, the authors confirm that the breeds cluster by phenotype or functional ability (Figure 3). They further refine the breed relationships by defining additional clusters within the established groupings such as scent hounds and spaniels within the fourth cluster, and sighthounds separate from herding dogs in the third cluster. This same data set was instrumental in mapping phenotypes that are specific to breed development. Recently, another study examining 1,375 dogs and 19 wolves at the same loci confirmed the separation of the Asian/African breeds from the modern breeds. The authors postulate that this division is likely the result of isolation of these breeds during the explosion of breed creation in Europe preventing hybridization between the breed types.65
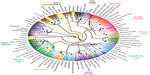  | Figure 3 Phylogenetic tree of 80 domestic dog breeds rooted with the coyote. |
Some of the earliest phenotype mapping in dogs was done through candidate gene studies of coat color, primarily in the MC1R/ASIP pathway (reviewed in Schmutz and Berryere66). These studies showed that most mutations affecting canine phenotypes were shared across breeds indicating that they arose once and were then maintained in the breeds where the result was desirable. However, in a minority of cases, multiple mutations could be found in the same gene with no breed specificity.67 For example, three mutations were identified in the TYRP1 gene that creates brown coat color. Combinations of these mutations were identified within the same breed and even within the same dog that suggest that the phenotype has arisen multiple times independently in the history of the dog, likely before the advent of the breeds.
The development of linkage and radiation hybrid maps of the dog genome allowed researchers to begin looking for novel gene variants that lead to major phenotypes. Using a linkage strategy in a crossbred pedigree of dogs, a new locus was identified that associated with solid black and brindle coat colors.68 Sequencing and haplotype analysis across multiple breeds found that the beta-defensin gene (CBD103), a gene usually associated with the immune system, was responsible for dominant black coat.69
The first GWAS performed in the dog analyzed very small numbers to identify trait-associated loci of large effect.70 Less than ten cases and ten controls were chosen to represent segregating coat types in Boxers and Rhodesian Ridgebacks and were genotyped at 27,000 SNPs. Complete sequencing of the associated haplotypes identified a 100 kb region around MITF as the source of white spotting in Boxer and Boxer-related breeds. A follow-up analysis found that a large CNV on CFA18 was responsible for the hair ridge in the Rhodesian Ridgebacks.71 The success of these association studies confirmed the power of the canine GWASs to uncover simple genetic traits.
Many of the most interesting traits found in dogs are those that define the breeds. These cannot be traced through pedigrees because they do not segregate within breeds (Figure 2). In order to identify these genes, large multi-breed mapping sets were developed. Brachycephaly, the foreshortened skull and snout, is a complex trait found in many breeds such as Bulldogs, Pugs, and Pekingese. Multi-breed GWASs have identified up to eight loci significantly associated with the skull phenotype.64,72,73 Homozygosity mapping and haplotype comparisons at one of these loci identified a missense mutation in BMP3 that was shown to have biological relevance through zebrafish knock-down and rescue assays.73 The combination of multi-breed GWAS and homozygosity/haplotype analysis has been used successfully to identify mutations responsible for traits such as leg length, coat type and color, skull shape, and body size.47,49,64,74–76
A comprehensive study of canine skeletal traits came from the collection of a large cohort of Portuguese Water Dogs (PWDs) with extensive morphologic data.77 Based on radiographic measurements, genome regions were associated with skeletal traits such as skull length, pelvic width, bone width, and overall body size. Body size was associated with two markers on chromosome 15, one of which was near the insulin-like growth factor 1 (IGF1). This finding was extended using a multi-breed approach to identify the causative alleles. The initial 4 Mb associated region was fine mapped by SNP genotyping both large and small PWDs as well as dogs from small (<9 kg) and large (>30 kg) breeds. A single haplotype was identified in all 14 small breeds that included the IGF1 gene. This same haplotype was present in the small PWDs clearly displaying that studies carried out in a single breed could be significant in other breeds, especially in the case of highly selected morphologic traits.
Variation in canine body size is a particularly popular subject in mapping studies.64,78,79 From the smallest breeds like the Chihuahua to the giant English Mastiff, there can be a tenfold increase in height and a 50-fold increase in weight.80 This variation is almost entirely inherited as it is maintained within breed structures where variation in size and weight is extremely low (Figure 4). A recent study shows that seven variants in six genes can explain 50% of size reduction in dogs.76 The smallest breeds carry all seven of the mutations, while breeds averaging 41 kg (90 lbs) or more rarely carry any of the mutations (Figure 5). In comparison, the mapping of height in humans has identified 180 loci and yet explains approximately 10% of observed variation.81 This is an excellent example of how canine population structure can help simplify the genetics of a complex phenotype. The constant selective pressure on desirable traits while maintaining current standards can fix the most effective mutations while eliminating the steady build of lesser deleterious mutations. If this axiom holds true across phenotypes, it will prove especially important when applied to disease mapping.
  | Figure 5 Alleles at six size loci combine to reduce size in dogs. |
The dog as a model to study human genetic disease
Dogs share more genetic similarity with humans than do traditional model organisms such as the mouse39 and an estimated 360 naturally occurring analogous diseases.82 Further, many of the traditional gene discovery methods discussed above (linkage analysis, GWAS) can be difficult and costly in humans due to the need for large numbers of samples to make up for short stretches of linkage disequilibrium and extensive disease heterogeneity. These same studies can be enhanced in dogs due to the predisposition to certain diseases within breeds where the unique population structure limits heterogeneity and increases linkage disequilibrium.82,83 As such, the dog offers an alternative and parallel system in which human diseases can be studied, both for discovering information about susceptibility and disease development and for predicting the course of a disease and optimal treatments.
Simple, monogenetic diseases in dogs often have the same genetic causes in humans (Table 1). One such example of this is progressive rod–cone degeneration (prcd) in dogs and its analogous disease in humans, retinitis pigmentosa (RP).84 In one of the first linkage-mapping studies, prcd was mapped to canine chromosome 9, orthologous to human chromosome 17q where a human RP locus was suggested to reside. Using haplotype analysis and a retinal cDNA library, a novel gene (PRCD) was identified that harbored a mutation segregating perfectly with the canine disease. The identical mutation was found in a woman diagnosed with autosomal recessive RP.85 This is a perfect example of how canine disease genetics mimics human and in this case allowed for the identification of a previously unknown gene and mutation. There are many other examples in which genetic mapping of diseases in dogs has led to the discovery of mutations in a homologous gene in humans, some of which can be found within the list of gene mutations in Table 1.
Single-gene disorders have played an important part in establishing the canine system as an exemplary counterpart to human studies; however, complex diseases are the area of greatest need. Because of the nature of the disorder, complex diseases are difficult to model in a laboratory as they require interactions or correlated action from multiple genes as well as some degree of environmental interaction. As an intermediate, the dogs provide an intriguing option to traditional models or human studies. They develop disease naturally through inheritance and interactions with our environment, yet their breed structure creates independent and unique strains in which heterozygosity is reduced and mapping is less complex.
One of the best examples of the canine role in elucidating genetic contributors to a complex human disorder can be found in the sleeping disorder narcolepsy. Canine narcolepsy with cataplexy was one of the first diseases mapped in the dog using microsatellites in a colony of narcoleptic Doberman Pinschers and Labrador Retrievers where the disorder segregated as an autosomal recessive trait. Extensive fine mapping and resequencing identified a SINE insertion in the HCRTR2 gene.86 This finding revealed a family of neurotransmitters that had not been previously associated with sleep. Shortly following this discovery, a mutation was identified in a human early-onset narcolepsy case, and more importantly, a deficiency in the hypocretin system was identified in the majority of cases of narcolepsy with cataplexy showing the importance of the gene family in the human disease and altering the future of sleep studies.87
Many of the most prevalent complex diseases in people are also very common in dogs such as diabetes, epilepsy, heart disease, and cancer. In 2005, a canine GWAS identified a repeat expansion in NHLRC1 (previously known as EPM2B) as the cause of a canine version of epilepsy in Dachshunds that is similar to Lafora disease.88 This finding coincided almost exactly with the discovery of mutations in the same gene in the human disease.89 Since then, mutations in ten different genes have been identified as causative for forms of epilepsy in ten different breeds of dog (reviewed in Ekenstedt and Oberbauer90). The availability of many breeds, each with independent disease inheritance, allows for the discovery of complex disease pathways, one gene at a time.
Cancers in dogs offer a unique opportunity to inform human disease. As a result of the founder effect, canine cancers are often breed specific or overrepresented in particular breeds. For instance, 25% of Bernese Mountain Dogs (BMDs) are estimated to develop histiocytic sarcoma, and 12.5% of Rottweilers develop osteosarcoma.91 That certain cancers tend to segregate within specific breeds reduces the genetic and environmental noise that is associated with those cancers, and allows for a genetic association to be made more readily than would be in human cases.
One example is histiocytic sarcoma, a rare, lethal cancer in humans that often strikes juveniles. It is also a rare tumor in dogs but is very common in BMDs allowing for mapping studies that can elucidate genetic factors that may be informative in both species. The first GWAS on histiocytic sarcoma was undertaken in BMD and revealed an association with the MTAP-CDKN2A locus on CFA11,92 a locus homologous to human 9p21 which is implicated in many human diseases including cancers.93 Similarly, squamous cell carcinoma of the digit in Standard Poodles, Giant Schnauzers, and Briards is associated with a CNV on CFA15 near KITLG.94 These studies capitalized on the minimal genetic heterozygosity of cancer inheritance in the dog. Lymphomas are common in both dogs and humans and represent an area of oncology study that would immediately benefit both species. A recent GWAS in Golden Retrievers has identified two loci on CFA5 that are associated with both lymphoma and hemangiosarcoma in the breed.95 Different forms of cancers associated with the same locus suggest that there are mutations that affect tumor formation in general and comparing different tumor types with the same causative mutations may help to ferret out the genetics behind tissue specificity.
Conclusion and future prospects
The promise of dog genetic studies lies in translation of our findings to improved treatments in both dogs and people. For instance, Golden Retriever muscular dystrophy, a homologue of Duchenne muscular dystrophy (DMD), is caused by exon skipping followed by early truncation of the dystrophin (dmd) gene, the same gene that is mutated in two thirds of human beings with DMD.96 Because dmd is one of the longest genes in the genome, new genetic approaches to treat DMD have been to promote in-frame exon skipping of the mutated area of dmd to produce a more functional, partial protein leading to a less severe phenotype. To find a tolerable and long-lasting therapy, a small nuclear RNA delivered through a recombinant adenovirus was tested on Golden Retrievers with muscular dystrophy.97,98 The success of these trials has recently led to a delivery and safety trial specifically staged to prepare for human trials.99
Muscular dystrophy is not the first area in which gene therapy has been used successfully in the dog leading to human trials. Multiple forms of canine progressive retinal atrophy have been successfully treated using gene supplementation (reviewed in Petersen-Jones100). Clinical trials on RPE65-deficient RP, Leber congenital amaurosis type 2, treated with gene augmentation therapy, showed stable vision improvement and no adverse effect over 3 years.101 Recent success in treating X-linked RP in the dog shows similar promise for translation to human.102
In addition to inherited mutations, studies are showing that canine diseases share many somatic alterations with human diseases. For example, the Philadelphia chromosome, a common translocation between human chromosomes 9 and 22 found in chronic myelogenous leukemia, is also found in dogs with the same disease (translocation between canine chromosomes 9 and 26).103 A similar translocation has been found in human and canine Burkitt lymphoma (between human chromosomes 8 and 14 and between canine chromosomes 13 and 8), and the RB1 gene locus is deleted in both human and canine chronic lymphocytic leukemia. These findings suggest that there are inherently fragile regions of the genome that support tumorigenesis and further comparison of human and canine tumor DNA may enable the discovery of novel cancer genes and mutations. Studies investigating osteosarcoma in dogs have suggested some candidate genes that may be important in the human disease. A comparison of the expression profiles of human and canine osteosarcoma found that overall, the expression profiles were indistinguishable, but a closer inspection of individual genes identified two genes that were expressed in all dogs but only a subset of humans. These genes, IL-8 and SLC1A3, are associated with a more aggressive form of the cancer.104
Highly similar mutation loads will allow for the testing of targeted treatments and therapies in dogs that have the potential to benefit humans as well. The goal of personalized medicine is to tailor treatment strategies to fit the individual patient. Sequencing of the transcriptome of canine invasive bladder cancers has recently identified the common human BRAFV600E mutations in >85% of tumors.105 Because the BRAFV600E mutations are present in 8% of all human cancers, they have been the focus of multiple clinical trials using targeted drug therapies.106 Identification of identical mutations in a naturally occurring dog tumor puts the dog at the forefront of preclinical trial strategies for highly specific therapies enabling the determination of dosage amounts, testing combination therapies, and determining efficacy prior to costly human trials.
Tremendous advances have been made in canine genetic and genomic studies since the first backcrosses were performed and proteins isolated. As a system for scientific discovery, canine genomic researchers now have the necessary tools and the ability to answer questions of inheritance, association, and causality. Novel treatment strategies have been introduced based on canine genetic studies, and the population structure encourages the development of assays to interrogate the effects of environmental and genetic background on inherited mutations. With modern molecular techniques, canine genomic information is booming, adding to our knowledge of the dog, how we have shaped its history, and how it, in turn, is helping us to improve our future.
Disclosure
The authors report no conflicts of interest associated with this work.
References
Germonpre M, Sablin MV, Stevens RE, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J Archaeol Sci. 2009;36(2):473–490. | |
Ovodov ND, Crockford SJ, Kuzmin YV, Higham TF, Hodgins GW, van der Plicht J. A 33,000-year-old incipient dog from the Altai Mountains of Siberia: evidence of the earliest domestication disrupted by the Last Glacial Maximum. PLoS One. 2011;6(7):e22821. | |
Thalmann O, Shapiro B, Cui P, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342(6160):871–874. | |
Druzhkova AS, Thalmann O, Trifonov VA, et al. Ancient DNA analysis affirms the canid from Altai as a primitive dog. PLoS One. 2013;8(3):e57754. | |
Drake AG, Coquerelle M, Colombeau G. 3D morphometric analysis of fossil canid skulls contradicts the suggested domestication of dogs during the late Paleolithic. Sci Rep. 2015;5:8299. | |
Tchernov E, Valla FF. Two new dogs, and other Natufian dogs, from the Southern Levant. J Archaeol Sci. 1997;24:65–95. | |
Trut L, Oskina I, Kharlamova A. Animal evolution during domestication: the domesticated fox as a model. Bioessays. 2009;31(3):349–360. | |
Vilà C, Savolainen P, Maldonado JE, et al. Multiple and ancient origins of the domestic dog. Science. 1997;276(5319):1687–1689. | |
Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002; 298(5598):1610–1613. | |
Pang JF, Kluetsch C, Zou XJ, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26(12):2849–2864. | |
Freedman AH, Gronau I, Schweizer RM, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10(1):e1004016. | |
Wang GD, Zhai W, Yang HC, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. | |
Kopaliani N, Shakarashvili M, Gurielidze Z, Qurkhuli T, Tarkhnishvili D. Gene flow between wolf and shepherd dog populations in Georgia (Caucasus). J Hered. 2014;105(3):345–353. | |
Sutter NB, Eberle MA, Parker HG, et al. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14(12):2388–2396. | |
Akey JM, Ruhe AL, Akey DT, et al. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci U S A. 2010;107(3):1160–1165. | |
Janecka J, Chowdhary B, Murphy W. Exploring the correlations between sequence evolution rate and phenotypic divergence across the Mammalian tree provides insights into adaptive evolution. J Biosci. 2012;37(5):897–909. | |
Axelsson E, Ratnakumar A, Arendt ML, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495(7441):360–364. | |
Coppinger R, Coppinger L. Dogs. New York: Scribner; 2001. | |
Santos JL, Saus E, Smalley SV, et al. Copy number polymorphism of the salivary amylase gene: implications in human nutrition research. J Nutrigenet Nutrigenomics. 2012;5(3):117–131. | |
Li Y, Wang GD, Wang MS, Irwin DM, Wu DD, Zhang YP. Domestication of the dog from the wolf was promoted by enhanced excitatory synaptic plasticity: a hypothesis. Genome Biol Evol. 2014; 6(11):3115–3121. | |
Gou X, Wang Z, Li N, et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24(8):1308–1315. | |
Storz JF, Moriyama H. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol. 2008;9(2):148–157. | |
Beall CM, Cavalleri GL, Deng L, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–11464. | |
Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–336. | |
Fuller JL. Genetic variability in some physiological constants of dogs. Am J Physiol. 1951;166(1):20–24. | |
Hare WC, Weber WT, McFeely RA, Yang TJ. Cytogenetics in the dog and cat. J Small Anim Pract. 1966;7(9):575–592. | |
Clough E, Pyle RL, Hare WC, Kelly DF, Patterson DF. An XXY sex-chromosome constitution in a dog with testicular hypoplasia and congenital heart disease. Cytogenetics. 1970;9(1):71–77. | |
Shive RJ, Hare WC, Patterson DF. Chromosome studies in dogs with congenital cardiac defects. Cytogenetics. 1965;4(6):340–348. | |
Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999;61(2):145–155. | |
Yang F, O’Brien PC, Milne BS, et al. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62(2):189–202. | |
Breen M. Canine cytogenetics – from band to basepair. Cytogenet Genome Res. 2008;120(1–2):50–60. | |
Cardenas JM, Wold F. Comparative studies on structural and catalytic properties of enolases. Arch Biochem Biophys. 1971;144(2):663–672. | |
Muto Y, Smith FR, Goodman DS. Comparative studies of retinol transport in plasma. J Lipid Res. 1973;14(5):525–532. | |
Jones GE. Comparative immunological studies between canine prolactin and prolactin from other species. Acta Endocrinol (Copenh). 1976;82(3):475–485. | |
Devilee P, Warnaar JN, Giphart MJ. MHC homology between various mammalian species at the DNA level: its relevance to MHC evolution. Exp Clin Immunogenet. 1984;1(2):90–98. | |
Storb R, Rudolph RH, Thomas ED. Marrow grafts between canine siblings matched by serotyping and mixed leukocyte culture. J Clin Invest. 1971;50(6):1272–1275. | |
Thomas ED, Bryant JI, Buckner CD, et al. Allogeneic marrow grafting using HL-A matched donor-recipient sibling pairs. Trans Assoc Am Physicians. 1971;84:248–261. | |
Storb R, Thomas ED. Graft-versus-host disease in dog and man: the Seattle experience. In: Moller G, editor. Immunological Reviews No 88. Copenhagen: Munksgaard; 1985:215–238. | |
Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. | |
Ostrander EA, Sprague GF Jr, Rine J. Identification and characterization of dinucleotide repeat (CA)n markers for genetic mapping in dog. Genomics. 1993;16(1):207–213. | |
Ostrander EA, Mapa FA, Yee M, Rine J. One hundred and one new simple sequence repeat-based markers for the canine genome. Mamm Genome. 1995;6(3):192–195. | |
Francisco LV, Langston AA, Mellersh CS, Neal CL, Ostrander EA. A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome. 1996;7(5):359–362. | |
Mellersh CS, Langston AA, Acland GM, et al. A linkage map of the canine genome. Genomics. 1997;46(3):326–336. | |
Priat C, Hitte C, Vignaux F, et al. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–378. | |
Wong AK, Ruhe AL, Dumont BL, et al. A comprehensive linkage map of the dog genome. Genetics. 2010;184(2):595–605. | |
Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9(9):713–725. | |
Vaysse A, Ratnakumar A, Derrien T, et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7(10):e1002316. | |
Forman OP, De Risio L, Matiasek K, Platt S, Mellersh C. Spinocerebellar ataxia in the Italian Spinone dog is associated with an intronic GAA repeat expansion in ITPR1. Mamm Genome. 2015;26(1–2):108–117. | |
Parker HG, VonHoldt BM, Quignon P, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325(5943):995–998. | |
Kirkness EF, Bafna V, Halpern AL, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301(5641):1898–1903. | |
Grall A, Guaguère E, Planchais S, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44(2):140–147. | |
Lavrijsen IC, Leegwater PA, Wangdee C, et al. Genome-wide survey indicates involvement of loci on canine chromosomes 7 and 31 in patellar luxation in Flat-Coated Retrievers. BMC Genet. 2014;15:64. | |
Agler C, Nielsen DM, Urkasemsin G, et al. Canine hereditary ataxia in old english sheepdogs and gordon setters is associated with a defect in the autophagy gene encoding RAB24. PLoS Genet. 2014;10(2):e1003991. | |
Guo J, O’Brien DP, Mhlanga-Mutangadura T, et al. A rare homozygous MFSD8 single-base-pair deletion and frameshift in the whole genome sequence of a Chinese Crested dog with neuronal ceroid lipofuscinosis. BMC Vet Res. 2015;10(1):960. | |
Downs LM, Mellersh CS. An Intronic SINE insertion in FAM161A that causes exon-skipping is associated with progressive retinal atrophy in Tibetan Spaniels and Tibetan Terriers. PLoS One. 2014;9(4):e93990. | |
Broeckx BJ, Coopman F, Verhoeven GE, et al. Development and performance of a targeted whole exome sequencing enrichment kit for the dog (Canis Familiaris Build 3.1). Sci Rep. 2014;4:5597. | |
Hoeppner MP, Lundquist A, Pirun M, et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One. 2014;9(3):e91172. | |
Zajc I, Sampson J. Utility of canine microsatellites in revealing the relationships of pure bred dogs. J Hered. 1999;90(1):104–107. | |
Koskinen MT, Bredbacka P. Assessment of the population structure of five Finnish dog breeds with microsatellites. Anim Genet. 2000;31(5):310–317. | |
Irion DN, Schaffer AL, Famula TR, Eggleston ML, Hughes SS, Pedersen NC. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J Hered. 2003;94(1):81–87. | |
Parker HG, Kim LV, Sutter NB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304(5674):1160–1164. | |
Parker HG, Kukekova AV, Akey DT, et al. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17(11):1562–1571. | |
Vonholdt BM, Pollinger JP, Lohmueller KE, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464(7290):898–902. | |
Boyko AR, Quignon P, Li L, et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8(8):e1000451. | |
Larson G, Karlsson EK, Perri A, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci U S A. 2012;109(23):8878–8883. | |
Schmutz SM, Berryere TG. Genes affecting coat colour and pattern in domestic dogs: a review. Anim Genet. 2007;38(6):539–549. | |
Schmutz SM, Berryere TG, Goldfinch AD. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm Genome. 2002;13(7):380–387. | |
Kerns JA, Cargill EJ, Clark LA, et al. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics. 2007;176(3):1679–1689. | |
Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276(8):5707–5713. | |
Karlsson EK, Baranowska I, Wade CM, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39(11):1321–1328. | |
Salmon Hillbertz NH, Isaksson M, Karlsson EK, et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39(11):1318–1320. | |
Bannasch D, Young A, Myers J, et al. Localization of Canine Brachycephaly using an across breed mapping approach. PLoS One. 2010;5(3):e9632. | |
Schoenebeck JJ, Hutchinson SA, Byers A, et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012;8(8):e1002849. | |
Cadieu E, Neff MW, Quignon P, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326(5949):150–153. | |
Dreger DL, Parker HG, Ostrander EA, Schmutz SM. Identification of a mutation that is associated with the saddle tan and black-and-tan phenotypes in Basset Hounds and Pembroke Welsh Corgis. J Hered. 2013;104(3):399–406. | |
Rimbault M, Beale HC, Schoenebeck JJ, et al. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 2013;23(12):1985–1995. | |
Chase K, Carrier DR, Adler FR, et al. Genetic basis for systems of skeletal quantitative traits: principal component analysis of the canid skeleton. Proc Natl Acad Sci U S A. 2002;99(15):9930–9935. | |
Hoopes BC, Rimbault M, Liebers D, Ostrander EA, Sutter NB. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm Genome. 2012;23(11–12):780–790. | |
Jones P, Chase K, Martin A, Davern P, Ostrander EA, Lark KG. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics. 2008;179(2):1033–1044. | |
Club AK. The Complete Dog Book. 19th ed. Garden City, NY: Doubleday; 1997. | |
Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. | |
Shearin AL, Ostrander EA. Leading the way: canine models of genomics and disease. Dis Model Mech. 2010;3(1–2):27–34. | |
Patterson D. Companion animal medicine in the age of medical genetics. J Vet Internal Med. 2000;14(1):1–9. | |
Acland GM, Ray K, Mellersh CS, et al. Linkage analysis and comparative mapping of canine progressive rod-cone degeneration (prcd) establishes potential locus homology with retinitis pigmentosa (RP17) in humans. Proc Natl Acad Sci U S A. 1998;96(6):3048–3053. | |
Zangerl B, Goldstein O, Philp AR, et al. Identical mutation in a novel retinal gene causes progressive rod-cone degeneration in dogs and retinitis pigmentosa in humans. Genomics. 2006;88(5):551–563. | |
Lin L, Faraco J, Li R, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. | |
Peyron C, Faraco J, Rogers W, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains [In Process Citation]. Nat Med. 2000;6(9):991–997. | |
Lohi H, Young EJ, Fitzmaurice SN, et al. Expanded repeat in canine epilepsy. Science. 2005;307(5706):81. | |
Gómez-Abad C, Gómez-Garre P, Gutiérrez-Delicado E, et al. Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology. 2005;64(6):982–986. | |
Ekenstedt KJ, Oberbauer AM. Inherited epilepsy in dogs. Top Companion Anim Med. 2013;28(2):51–58. | |
Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci. 2013;2013:941275. | |
Shearin AL, Hedan B, Cadieu E, et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1019–1027. | |
Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11(5):727–731. | |
Karyadi DM, Karlins E, Decker B, et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 2013;9(3):e1003409. | |
Tonomura N, Elvers I, Thomas R, et al. Genome-wide association study identifies shared risk loci common to two malignancies in Golden Retrievers. PLoS Genet. 2015;11(2):e1004922. | |
Sharp NJ, Kornegay JN, Van Camp SD, et al. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics. 1992;13(1):115–121. | |
Vulin A, Barthélémy I, Goyenvalle A, et al. Muscle function recovery in golden retriever muscular dystrophy after AAV1-U7 exon skipping. Mol Ther. 2012;20(11):2120–2133. | |
Bish LT, Sleeper MM, Forbes SC, et al. Long-term restoration of cardiac dystrophin expression in golden retriever muscular dystrophy following rAAV6-mediated exon skipping. Mol Ther. 2012;20(3):580–589. | |
Le Guiner C, Montus M, Servais L, et al. Forelimb treatment in a large cohort of dystrophic dogs supports delivery of a recombinant AAV for exon skipping in Duchenne patients. Mol Ther. 2014;22(11):1923–1935. | |
Petersen-Jones SM. Viral vectors for targeting the canine retina: a review. Vet Ophthalmol. 2012;15(Suppl 2):29–34. | |
Testa F, Maguire AM, Rossi S, et al. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital amaurosis type 2. Ophthalmol. 2013;120(6):1283–1291. | |
Beltran WA, Cideciyan AV, Lewin AS, et al. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci U S A. 2012;109(6):2132–2137. | |
Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans – man and his best friend share more than companionship. Chromosome Res. 2008;16(1):145–154. | |
Paoloni M, Davis S, Lana S, et al. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. | |
Decker B, Parker HG, Dhawan D, et al. Homologous mutation to human BRAF V600E is common in naturally occurring canine bladder cancer-evidence for a relevant model system and urine-based diagnostic test. Mol Cancer Res. Epub 2015 Mar 12. | |
Martin-Liberal J, Larkin J. New RAF kinase inhibitors in cancer therapy. Expert Opin Pharmacother. 2014;15(9):1235–1245. | |
Zheng K, Perry J, Harvey SJ, et al. Regulation of collagen type IV genes is organ-specific: evidence from a canine model of Alport syndrome. Kidney Int. 2005;68(5):2121–2130. | |
Winkelstein JA, Cork LC, Griffin DE, Griffin JW, Adams RJ, Price DL. Genetically determined deficiency of the third component of complement in the dog. Science. 1981;212(4499):1169–1170. | |
Alper CA, Propp RP. Genetic polymorphism of the third component of human complement (C’3). J Clin Invest. 1968;47(9):2181–2191. | |
Pele M, Tiret L, Kessler JL, Blot S, Panthier JJ. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14(11):1417–1427. | |
Muhammad E, Reish O, Ohno Y, et al. Congenital myopathy is caused by mutation of HACD1. Hum Mol Genet. 2013;22(25):5229–5236. | |
Böhm J, Vasli N, Maurer M, et al. Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 2013;9(6):e1003430. | |
Wolf ZT, Brand HA, Shaffer JR, et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 2015;11(3):e1005059. | |
Sidjanin DJ, Lowe JK, McElwee JL, et al. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11(16):1823–1833. | |
Kohl S, Baumann B, Broghammer M, et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet. 2000;9(14):2107–2116. | |
Henthorn PS, Liu J, Gidalevich T, et al. Canine cystinuria: polymorphism in the canine SLC3A1 gene and identification of a nonsense mutation in cystinuric Newfoundland dogs. Hum Genet. 2000;107(4):295–303. | |
Calonge MJ, Gasparini P, Chillarón J, et al. Cystinuria caused by mutations in rBAT, a gene involved in the transport of cystine. Nat Genet. 1994;6(4):420–425. | |
Awano T, Johnson GS, Wade CM, et al. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106(8):2794–2799. | |
Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. | |
Cooper BJ, Winand NJ, Stedman H, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–156. | |
Baldeschi C, Gache Y, Rattenholl A, et al. Genetic correction of canine dystrophic epidermolysis bullosa mediated by retroviral vectors. Hum Mol Genet. 2003;12(15):1897–1905. | |
Ryynanen M, Knowlton RG, Parente MG, Chung LC, Chu ML, Uitto J. Human type VII collagen: genetic linkage of the gene (COL7A1) on chromosome 3 to dominant dystrophic epidermolysis bullosa. Am J Hum Genet. 1991;49(4):797–803. | |
Whitney KM, Goodman SA, Bailey EM, Lothrop CD Jr. The molecular basis of canine pyruvate kinase deficiency. Exp Hematol. 1994;22(9):866–874. | |
Valentine WN, Tanaka KR, Miwa S. A specific erythrocyte glycolytic enzyme defect (pyruvate kinase) in three subjects with congenital non-spherocytic hemolytic anemia. Trans Assoc Am Physicians. 1961;74:100–110. | |
Skelly BJ, Sargan DR, Herrtage ME, Winchester BG. The molecular defect underlying canine fucosidosis. J Med Genet. 1996;33(4):284–288. | |
Willems PJ, Darby JK, DiCioccio RA, et al. Identification of a mutation in the structural alpha-L-fucosidase gene in fucosidosis. Am J Hum Genet. 1988;43(5):756–763. | |
Kishnani PS, Faulkner E, VanCamp S, et al. Canine model and genomic structural organization of glycogen storage disease type Ia (GSD Ia). Vet Pathol. 2001;38(1):83–91. | |
Lei KJ, Pan CJ, Shelly LL, Liu JL, Chou JY. Identification of mutations in the gene for glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1a. J Clin Invest. 1994;93(5):1994–1999. | |
Lozier JN, Dutra A, Pak E, et al. The Chapel Hill hemophilia A dog colony exhibits a factor VIII gene inversion. Proc Natl Acad Sci U S A. 2002;99(20):12991–12996. | |
Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5(3):236–241. | |
Mauser AE, Whitlark J, Whitney KM, Lothrop CD Jr. A deletion mutation causes hemophilia B in Lhasa Apso dogs. Blood. 1996;88(9):3451–3455. | |
Giannelli F, Choo KH, Rees DJ, Boyd Y, Rizza CR, Brownlee GG. Gene deletions in patients with haemophilia B and anti-factor IX antibodies. Nature. 1983;303(5913):181–182. | |
Mellersh CS, Graves KT, McLaughlin B, et al. Mutation in HSF4 associated with early but not late-onset hereditary cataract in the Boston Terrier. J Hered. 2007;98(5):531–533. | |
Bu L, Jin Y, Shi Y, et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31(3):276–278. | |
Lingaas F, Comstock KE, Kirkness EF, et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum Mol Genet. 2003;12(23):3043–3053. | |
Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2(2):157. | |
Casal ML, Scheidt JL, Rhodes JL, Henthorn PS, Werner P. Mutation identification in a canine model of X-linked ectodermal dysplasia. Mamm Genome. 2005;16(7):524–531. | |
Kere J, Srivastava AK, Montonen O, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13(4):409–416. | |
Owczarek-Lipska M, Jagannathan V, Drögemüller C, et al. A frameshift mutation in the cubilin gene (CUBN) in Border Collies with Imerslund-Grasbeck syndrome (selective cobalamin malabsorption). PLoS One. 2013;8(4):e61144. | |
Aminoff M, Carter JE, Chadwick RB, et al. Mutations in CUBN, encoding the intrinsic factor-vitamin B12 receptor, cubilin, cause hereditary megaloblastic anaemia 1. Nat Genet. 1999;21(3):309–313. | |
Kennedy LJ, Quarmby S, Happ GM, et al. Association of canine hypothyroidism with a common major histocompatibility complex DLA class II allele. Tissue Antigens. 2006;68(1):82–86. | |
Owerbach D, Gunn S, Ty G, Wible L, Gabbay KH. Oligonucleotide probes for HLA-DQA and DQB genes define susceptibility to type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1988;31(10):751–757. | |
Chan EM, Bulman DE, Paterson AD, et al. Genetic mapping of a new Lafora progressive myoclonus epilepsy locus (EPM2B) on 6p22. J Med Genet. 2003;40(9):671–675. | |
Hungs M, Fan J, Lin L, Lin X, Maki RA, Mignot E. Identification and functional analysis of mutations in the hypocretin (orexin) genes of narcoleptic canines. Genome Res. 2001;11(4):531–539. | |
Mole S. Neuronal ceroid lipofuscinoses. Eur J Paediatr Neurol. 2001;5(5):211–212. | |
Gardiner RM. The molecular genetic basis of the neuronal ceroid lipofuscinoses. Neurol Sci. 2000;21(3 Suppl):S15–S19. | |
Capt A, Spirito F, Guaguere E, Spadafora A, Ortonne JP, Meneguzzi G. Inherited junctional epidermolysis bullosa in the German pointer: establishment of a large animal model. J Invest Dermatol. 2005;124(3):530–535. | |
Kivirikko S, McGrath JA, Baudoin C, et al. A homozygous nonsense mutation in the alpha 3 chain gene of laminin 5 (LAMA3) in lethal (Herlitz) junctional epidermolysis bullosa. Hum Mol Genet. 1995;4(5):959–962. | |
Victoria T, Rafi MA, Wenger DA. Cloning of the canine GALC cDNA and identification of the mutation causing globoid cell leukodystrophy in West Highland White and Cairn terriers. Genomics. 1996;33(3):457–462. | |
Sakai N, Inui K, Fujii N, et al. Krabbe disease: isolation and characterization of a full-length cDNA for human galactocerebrosidase. Biochem Biophys Res Commun. 1994;198(2):485–491. | |
Kijas JM, Bauer TR Jr, Gafvert S, et al. A missense mutation in the beta-2 integrin gene (ITGB2) causes canine leukocyte adhesion deficiency. Genomics. 1999;61(1):101–107. | |
Kishimoto TK, Hollander N, Roberts TM, Anderson DC, Springer TA. Heterogeneous mutations in the beta subunit common to the LFA-1, Mac-1, and p150,95 glycoproteins cause leukocyte adhesion deficiency. Cell. 1987;50(2):193–202. | |
Rhodes TH, Vite CH, Giger U, Patterson DF, Fahlke C, George AL Jr. A missense mutation in canine C1C-1 causes recessive myotonia congenita in the dog. FEBS Lett. 1999;456(1):54–58. | |
Zhang J, George AL Jr, Griggs RC, et al. Mutations in the human skeletal muscle chloride channel gene (CLCN1) associated with dominant and recessive myotonia congenita. Neurology. 1996;47(4):993–998. | |
Aguirre GD, Baldwin V, Pearce-Kelling S, Narfstrom K, Ray K, Acland GM. Congenital stationary night blindness in the dog: common mutation in the RPE65 gene indicates founder effect. Mol Vis. 1998;4:23. | |
Marlhens F, Bareil C, Griffoin JM, et al. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat Genet. 1997;17(2):139–141. | |
Petersen-Jones SM, Entz DD, Sargan DR. cGMP phosphodiesterase-alpha mutation causes progressive retinal atrophy in the Cardigan Welsh corgi dog. Invest Ophthalmol Vis Sci. 1999;40(8):1637–1644. | |
Dryja TP, Finn JT, Peng YW, McGee TL, Berson EL, Yau KW. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A. 1995;92(22):10177–10181. | |
Kijas JW, Cideciyan AV, Aleman TS, et al. Naturally-occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 2002;99:6328–6333. | |
Bunge S, Wedemann H, David D, et al. Molecular analysis and genetic mapping of the rhodopsin gene in families with autosomal dominant retinitis pigmentosa. Genomics. 1993;17(1):230–233. | |
Kukekova AV, Goldstein O, Johnson JL, et al. Canine RD3 mutation establishes rod-cone dysplasia type 2 (rcd2) as ortholog of human and murine rd3. Mamm Genome. 2009;20(2):109–123. | |
Friedman JS, Chang B, Kannabiran C, et al. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am J Hum Genet. 2006;79(6):1059–1070. | |
Henthorn PS, Somberg RL, Fimiani VM, Puck JM, Patterson DF, Felsburg PJ. IL-2R gamma gene microdeletion demonstrates that canine X-linked severe combined immunodeficiency is a homologue of the human disease. Genomics. 1994;23:69–74. | |
Puck JM, Deschênes SM, Porter JC, et al. The interleukin-2 receptor gamma chain maps to Xq13.1 and is mutated in X-linked severe combined immunodeficiency, SCIDX1. Hum Mol Genet. 1993;2(8):1099–1104. | |
van de Leemput J, Chandran J, Knight MA, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3(6):e108. | |
Boudreaux MK, Catalfamo JL. Molecular and genetic basis for thrombasthenic thrombopathia in otterhounds. Am J Vet Res. 2001;62(11):1797–1804. | |
Kannan M, Ahmad F, Yadav BK, Kumar R, Choudhry VP, Saxena R. Molecular defects in ITGA2B and ITGB3 genes in patients with Glanzmann thrombasthenia. J Thromb Haemost. 2009;7(11):1878–1885. | |
Brooks M. A review of canine inherited bleeding disorders: biochemical and molecular strategies for disease characterization and carrier detection. J Hered. 1999;90(1):112–118. | |
Chen L, Deng CX. Roles of FGF signaling in skeletal development and human genetic diseases. Front Biosci. 2005;10:1961–1976. | |
van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet. 2002;11(2):165–173. | |
Tao TY, Liu F, Klomp L, Wijmenga C, Gitlin JD. The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J Biol Chem. 2003;278(43):41593–41596. | |
Horwitz M, Benson KF, Duan Z, Li FQ, Person RE. Hereditary neutropenia: dogs explain human neutrophil elastase mutations. Trends Mol Med. 2004;10(4):163–170. | |
Chen X, Johnson GS, Schnabel RD, et al. A neonatal encephalopathy with seizures in standard poodle dogs with a missense mutation in the canine ortholog of ATF2. Neurogenetics. 2008;9(1):41–49. | |
Raivich G, Behrens A. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 2006;78(6):347–363. | |
Seppälä EH, Jokinen TS, Fukata M, et al. LGI2 truncation causes a remitting focal epilepsy in dogs. PLoS Genet. 2011;7(7):e1002194. | |
Morante-Redolat JM, Gorostidi-Pagola A, Piquer-Sirerol S, et al. Mutations in the LGI1/Epitempin gene on 10q24 cause autosomal dominant lateral temporal epilepsy. Hum Mol Genet. 2002;11(9):1119–1128. | |
Battey J, Moulding C, Taub R, et al. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983;34(3):779–787. | |
Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16(2):109–112. | |
Boulalas I, Zaravinos A, Delakas D, Spandidos DA. Mutational analysis of the BRAF gene in transitional cell carcinoma of the bladder. Int J Biol Markers. 2009;24(1):17–21. | |
Tang R, Noh HJ, Wang D, et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 2014;15(3):R25. | |
Moya PR, Dodman NH, Timpano KR, et al. Rare missense neuronal cadherin gene (CDH2) variants in specific obsessive-compulsive disorder and Tourette disorder phenotypes. Eur J Hum Genet. 2013; 21(8):850–854. | |
Hejjas K, Vas J, Topal J, et al. Association of polymorphisms in the dopamine D4 receptor gene and the activity-impulsivity endophenotype in dogs. Anim Genet. 2007;38(6):629–633. | |
Faraone SV, Biederman J, Weiffenbach B, et al. Dopamine D4 gene 7-repeat allele and attention deficit hyperactivity disorder. Am J Psychiatry. 1999;156(5):768–770. | |
Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. 5th ed. Neptune City, NJ: TFH Publications; 1995. | |
Dog Genome Browser Gateway. University of California, Santa Cruz Genome Browser website. Available from: genome.ucsc.edu/cgi-bin/hgGateway?org=Dog. Accessed March 24, 2015. | |
Dog Genome Assembly. Ensembl Genome browser website. Available from: www.ensembl.org/Canis_familiaris/Info/Index?db=core. Accessed March 24, 2015. | |
Canis lupus familiaris (dog) Genome. National Center for Biotechnology Information website. Available from: www.ncbi.nlm.nih.gov/genome/?term=dog. Accessed March 24, 2015. | |
Broad improved canine annotation v1. Track Data Hubs. University of California, Santa Cruz Genome Browser website. Available from: genome.ucsc.edu/cgi-bin/hgHubConnect. Accessed March 24, 2015. | |
Dog gene expression. Ensembl Genome browser website. Available from: www.ensembl.org/Canis_familiaris/Info/Expression?db=core. Accessed March 24, 2015. | |
CHORI-82: Canine Boxer (F) (Canis familiaris) BAC Library. Children’s Hospital Oakland Research Institute website. Available from: bacpac.chori.org/library.php?id=253. Accessed March 24, 2015. | |
RPCI-81 Male (Doberman Pinscher) Canine BAC Library. Children’s Hospital Oakland Research Institute website. Available from: bacpac.chori.org/mcanine81.htm. Accessed March 24, 2015. | |
CanineHD Whole-Genome Genotyping. Illumina, Inc. website. Available from: illumina.com/products/caninehd_whole_genome_genotyping.html. Accessed March 24, 2015. | |
Canine SNP Genotyping Array. Affymetrix, Inc. website. Available from: www.affymetrix.com/estore/catalog/prod670005/AFFY/Canine. Accessed March 24, 2015. | |
Canine Array FAQ. Broad Institute website. Available from: www.broadinstitute.org/mammals/dog/caninearrayfaq.html. Accessed March 24, 2015. | |
Agilent Canine Oligo Microarray. Agilent Technologies website. Available from: www.chem.agilent.com/Library/brochures/5989-4567eN_72.pdf. Accessed March 24, 2015. | |
Canine array comparative genomic hybridization (aCGH). Breen Lab at North Carolina State University website. Available from: www.breenlab.org/array(new).html. Accessed March 24, 2015. | |
Model Organism CGH Microarrays – Details & Specifications. Agilent Technologies website. Available from: www.genomics.agilent.com/article.jsp?pageId=1544. Accessed March 24, 2015. | |
Past Versions of the Canine Genome Map. National Human Genome Research Institute Dog Genome Project website. Available from: research.nhgri.nih.gov/dog_genome/map_history/index.shtml. Accessed March 24, 2015. | |
Canine Genetic Linkage Map. Veterinary Genetics Laboratory at the University of California, Davis website. Available from: www.vgl.ucdavis.edu/dogmap/. Accessed March 24, 2015. | |
A 1 Mb Resolution Radiation Hybrid Map of the Canine Genome. National Human Genome Research Institute Dog Genome Project website. Available from: research.nhgri.nih.gov/dog_genome/guyon2003/index.shtml. Accessed March 24, 2015. | |
AutoGRAPH: A Comparative Genomics Server. AutoGRAPH website. Available from: autograph.genouest.org/index_pre.php. Accessed March 24, 2015. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




