Back to Archived Journals » Cell Health and Cytoskeleton » Volume 8
Expression profile of plakin cross-linking proteins in short-term denervated mouse hindlimb skeletal muscle
Authors Blouin P, Serresse O, Dorman S, Larivière C
Received 27 April 2016
Accepted for publication 28 June 2016
Published 18 August 2016 Volume 2016:8 Pages 37—46
DOI https://doi.org/10.2147/CHC.S111482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Denis Wirtz
Patrick Blouin,1 Olivier Serresse,1 Sandra C Dorman,1,2 Céline Larivière,1–3
1School of Human Kinetics, 2Northern Ontario School of Medicine, 3Biomolecular Sciences, Laurentian University, Sudbury, ON, Canada
Purpose: Skeletal muscle atrophy linked to neuromuscular inactivity is a complex phenomenon involving widespread alteration of muscle structure and function. Plakin cross-linking proteins are important structural elements that are expressed in skeletal muscles, which require resistance to mechanical stress. The plakin proteins most prevalent in skeletal muscles are plectin, dystonin, and microtubule-actin cross-linking factor (MACF). The expression profile of plakin cross-linking proteins in skeletal muscles during atrophy is poorly understood. We therefore investigated the expression profile of plectin, dystonin, and MACF in mouse hindlimb muscles undergoing denervation-induced atrophy.
Materials and methods: Quantitative polymerase chain reaction and Western blotting were performed to assess plakin mRNA and protein expression in mouse gastrocnemius muscles that were denervated for 1 day, 3 days, 7 days, and 14 days. The left hindlimb muscles were denervated by severing the left sciatic nerve, and the contralateral limb muscles served as sham control muscles. The mRNA expression of myogenin and acetylcholinesterase was measured in parallel and served as controls.
Results: As expected, myogenin mRNA expression was substantially induced in denervated muscles (13-fold induction), whereas acetylcholinesterase expression was significantly and progressively suppressed (90% reduction) in denervated skeletal muscles. In comparison, we found that plectin and dystonin mRNA expression were progressively reduced by ~50% at day 14 denervation time point, but the protein levels remained relatively constant. On the other hand, MACF expression was upregulated approximately threefold by day 7 denervation at both the mRNA and protein levels.
Conclusion: This study suggests that plakins may be of potential importance in the baseline preservation of the skeletal muscle structure following short-term denervation-induced atrophy. These results shed light on the gene expression profile of the skeletal muscle plakins during short-term denervation-induced atrophy.
Keywords: dystonin, plectin, MACF, muscle atrophy
Introduction
The structural organization of skeletal muscle fibers ensures that the fibers are able to withstand significant mechanical stress associated with cycles of muscle contraction and relaxation.1 Proteins belonging to the plakin family are believed to act as scaffolding proteins to solidify the cytoarchitecture of muscle fibers and other cell types.2–4 Plectin, dystonin (also known as Bpag1), and microtubule-actin cross-linking factor (MACF) are plakin family members known to be expressed in skeletal muscles.5–7 Expression deficiencies in the plectin, dystonin, and MACF genes in rodents can have effects ranging from embryonic lethality (MACF null) to severe skin blistering and death shortly after birth (plectin null) and death 2–3 weeks after birth (dystonin deficiency).7–13 MACF is ubiquitously expressed during embryonic development but is barely detectable in adult skeletal muscles.6 Thus, the role of MACF, if any, in adult skeletal muscles is currently not known. Plectin and dystonin are believed to bind and stabilize sarcomeres at the z-disk7,14,15 and to associate with mitochondria via interactions with desmin.5,16
Muscle atrophy resulting from neuromuscular inactivation is a complex molecular process.17–19 Atrophy can be induced experimentally through hindlimb suspension, hindlimb immobilization, chemical denervation (botulinum toxin), and surgical denervation (severing the nerve).20–24 All these methods share the common trait of reducing or completely inhibiting neural input to the muscle, which induces atrophy. Denervation of adult muscles can revert the expression of specific genes to an embryonic expression profile. For instance, γ-subunits of the Nicotinic acetylcholine receptor (nAChR) as well as myogenin, which are highly expressed in embryonic muscles and much less expressed in adult muscles, are reexpressed following denervation.25–27 In addition, many cytoskeletal proteins, such as desmin, tubulin, titin, and elements of the dystroglycan complex such as dystrophin and β-dystroglycan, are differentially regulated following denervation.28–31 There is currently sparse information regarding the expression patterns of plectin, dystonin, and MACF in muscles during denervation-induced atrophy. We therefore assessed the expression of plectin, dystonin, and MACF in denervated mouse hindlimb muscles after 1 day, 3 days, 7 days, and 14 days of denervation to shed light on the expression pattern of these proteins during early-onset atrophy. We found MACF upregulation at the protein and mRNA levels, while plectin and dystonin were downregulated at the mRNA level and their expression was stable at the protein level.
Materials and methods
Mouse information and denervation procedure
Two-month-old male CD1 mice were purchased from Charles River Laboratories (Saint-Constant, QC, Canada) and were kept in plastic shoebox cages in groups of four to five mice per cage. Mice had access to food and water ad libitum and were exposed to a 12-hour light/dark cycle in a climate-controlled room (23±2°C). Animals were treated in accordance with the Canadian Council on Animal Care guidelines. This study was approved by the Laurentian University Animal Care Committee.
Mice were anesthetized using isoflurane and were denervated unilaterally by severing the sciatic nerve near the hip area. The opposite limb was sham-denervated: these limbs had their sciatic nerve exposed, but it was not severed. Mice were then sutured with sterile clips and left to recover. Mice behavior was monitored following surgery and for the length of the experimental period. Oral analgesics were administered regularly to the animals within the first 24 hours postsurgery. None of the mice displayed signs of pain (ie, lack of grooming and low food intake). The denervation status was confirmed by the absence of the toe pinch, toe spread, and plantar flexion reflexes. After 1 day, 3 days, 7 days, or 14 days, mice were anesthetized with a xylazine (10 mg/kg bodyweight) and ketamine HCl (100 mg/kg bodyweight) mix, and the gastrocnemius skeletal muscles were carefully harvested from the denervated and sham hindlimb and quickly frozen in liquid nitrogen. Muscle samples were stored at -80°C.
Sample preparation
Mice gastrocnemius muscles were divided into two halves; one half was processed for RNA extraction and the other half for protein isolation. Before being divided, muscles were weighed whole while still frozen and their total dry weight was recorded. For RNA isolation, TRIzol® (Thermo Fisher Scientific, Waltham, MA, USA) was used as per the manufacturer’s instructions. RNA pellets were resuspended in RNase-free water and then frozen at -80°C until further use. For protein isolation, muscle samples were homogenized in radioimmune precipitation assay (RIPA) buffer supplemented with protease inhibitors (RIPA: 150 mM sodium chloride, 1.0%, v/v, Triton X-100, 0.5%, w/v, sodium deoxycholate, 0.1%, w/v, sodium dodecyl sulfate, 50 mM Tris, pH 8.0; protease inhibitors: 2 μg/mL aprotinin, 10 μg/mL leupeptin, 1 μg/mL Pepstatin A, 1 mM PMSF, 5 mM EDTA, 1 mM EGTA, 10 mM sodium fluoride, and 1 mM sodium orthovanadate). RIPA buffer (300 μL) was used per 25 mg of tissue. To homogenize the muscle, a Tissue Tearor (BioSpec products, Bartsville, OK, USA) was used in 15-second pulses until samples were homogenous. Samples were then incubated at 4°C for 4 hours under constant agitation and then centrifuged (12,000 rpm at 4°C) for 20 minutes. The supernatant from each sample was collected and stored at -80°C until further use. RNA and protein concentrations for each sample were then determined using standard methods.
Immunoblot analysis
Eight protein samples obtained from sham and denervated gastrocnemius muscles at each time point (1 day, 3 days, 7 days, and 14 days) were treated with Laemmli’s buffer (63 mM Tris HCl, 10%, v/v, glycerol, 2%, w/v, SDS, 0.0025%, w/v, bromophenol blue, pH 6.8, 5% b-mercaptoethanol, v/v) and were heated at 100°C for 5 minutes and then cooled for 5 minutes on ice prior to loading on an SDS/4% polyacrylamide gel (100 mg total protein per sample; 4% stacking, pH 6.8; 4% resolving, pH 8.8) along with a sample of HiMark pre-stained protein ladder (LC5699; Thermo Fisher Scientific). Samples were electrophoresed for 120 minutes in SDS electrophoresis buffer (25 mM Tris base, 190 mM glycine, and 0.1%, w/v, SDS) and were then transferred onto a polyvinylidene fluoride membrane (0.45 μM pore size; EMD Millipore, Billerica, MA, USA) in transfer buffer (25 mM Tris base, 190 mM glycine, and 15%, v/v, methanol). Membranes were then blocked for 1 hour in 5% (w/v) milk in Tween Tris-buffered saline (TTBS) (50 mM Tris, 150 mM NaCl, 0.05%, v/v, Tween 20, and pH 7.6) and then incubated with either mouse monoclonal plectin (sc-33649; Santa Cruz Biotechnology Inc., Dallas, TX, USA), polyclonal rabbit dystonin (custom; Sigma-Aldrich Co., St Louis, MO, USA), or mouse monoclonal MACF (sc-68430; Santa Cruz Biotechnology Inc.) antibody at 1:200 dilution in 5% milk in TTBS overnight at 4°C under constant agitation. Membranes were then washed in three separate 10-minute TTBS washes and then incubated with the corresponding goat antimouse (sc-2005; Santa Cruz Biotechnology Inc.) or antirabbit (sc-2030; Santa Cruz Biotechnology Inc.) secondary antibody conjugated to horseradish peroxidase at a dilution of 1:50,000 in 5% milk in TTBS at room temperature for 30 minutes on a rocking platform (Model 200; VWR Scientific Mississauga, Ontario, Canada). The membranes were washed again (3×10-minute washes in TTBS) and subsequently developed with enhanced chemiluminescence reagents (RPN2232; GE Healthcare UK Ltd, Little Chalfont, UK). Then, the membranes were stripped of antibodies, reprobed after each immunoblotting experiment, and stripped a final time before staining the membrane with Coomassie brilliant blue to verify protein loading to be used as a loading control. After Coomassie staining the membranes, they were imaged using the ChemiDoc XRS machine (Bio-Rad Laboratories Inc., Hercules, CA, USA). Densitometry analyses were performed by outlining the bands and subtracting the background intensity from the average band intensity using the Quantity One software (Bio-Rad Laboratories Inc.).
Quantitative polymerase chain reaction analysis
RNA was first reverse transcribed using the Qiagen QuantiTect Reverse Transcription kit, as per manufacturer’s instructions. One microgram of total RNA was used for the reverse transcription reaction in a total volume of 20 µL. For polymerase chain reaction (PCR) analysis, the Qiagen QuantiTect Primer Assay was used with the following primers: actin (QT00095242), plectin (QT00288589), myogenin (QT00112378), acetylcholinesterase (AChE; QT00287357), dystonin (QT01771308), tubulin (QT00116319), MACF (QT00243670), and ribosomal protein S12 (QT00317317) as per manufacturer’s instructions. We used 100 ng of total cDNA in a 25 µL total reaction volume for the PCR. Eight denervated samples from each (1 day, 3 days, 7 days, and 14 days) time point, along with the corresponding contralateral sham control sample, were loaded in duplicate onto 96-well plates, incubated at 95°C for 15 minutes followed by 40 cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds in a PTC-200 thermal cycler equipped with a Chromo4 Real-Time PCR instrument (Bio-Rad Laboratories Inc.). PCR data were normalized to ribosomal protein S12 as it was considered to be the most reliable internal control (ie, stably expressed in sham vs denervated [den] samples) (Figure 1). Fold change was calculated using the 2(–ΔΔCt) method. The average quantitative PCR (qPCR) efficiency was 92%.
Data analysis
Data were analyzed using SPSS. Outliers were removed according to default SPSS settings for outliers, defined as 1.5 times the interquartile (Q3–Q1) range below the first and above the third quartile. Data were checked for normality and when the test for normality failed, the data were log-transformed and rechecked for normality. When this improved normality, log-transformed data were used for analysis. This was the case for MACF mRNA and protein data. An analysis of variance (ANOVA) was used to determine differences between den/sham ratios across the four time points for each of eight data sets (plectin, dystonin, MACF, myogenin, AChE mRNA, plectin, dystonin, and MACF protein expression), and paired t-tests were used to assess the difference between den and sham pairs within each time point for each of the protein data sets above. The significance threshold was set at P<0.05.
Results
Markers of denervation
Frozen mouse gastrocnemius muscles were weighed, and these values were normalized to total bodyweight of each mouse (milligram of muscle per gram of bodyweight). These values are reported in Table 1. As anticipated, significant muscle atrophy was observed in denervated hindlimb muscles as revealed by paired t-tests. In particular, normalized muscle mass had decreased by 6.6% after only 24 hours of denervation (P=0.006), with a 41.5% reduction after 14 days of denervation (P<0.001). In comparison, the left and right gastrocnemius muscles of control mice, which were anesthetized but did not undergo surgery, had nearly identical muscle mass (P=1.000), thereby indicating that our excision technique was consistent.
Next, we measured the gene expression levels of myogenin and AChE in sham vs den skeletal muscle at the same four denervation time points using qPCR. Myogenin mRNA levels were upregulated 16-fold in day 1 denervated muscle, 24-fold at day 3 and day 7, and sevenfold at day 14. ANOVA results comparing the fold increase between 1 day and 2 weeks of denervation revealed no significant differences between fold increases at the various time points (F3, 28=2.220, P=0.108). In contrast, AChE was downregulated, as expected, by ~75% after 1 day of inactivation, by 87% after 3 days, 65% after 7 days, and 95% after 14 days. In this instance, the ANOVA was significant (F3, 28=7.468, P=0.001) with day 14 expression levels being significantly lower than day 7 expression levels. Collectively, the muscle weight and myogenin/AChE gene expression profiles are in line with the anticipated phenotype of denervated skeletal muscle. We, therefore, proceeded to measure the expression profile of the three plakin family members both at the mRNA and protein levels to better understand how the expression patterns of these cytoskeletal proteins are impacted in skeletal muscles that have been inactivated subsequent to motor neuron injury.
mRNA expression of plectin, dystonin, and MACF in denervated skeletal muscle
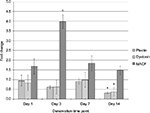
Using the same cDNA samples processed for qPCR experiments to measure myogenin and AChE expressions, we determined the gene expression levels of plectin, dystonin, and MACF. These results are presented in Figure 2. For plectin, the mRNA levels were reduced by 68% in day 14 denervated muscle, and this level was significantly lower than after 1 day of inactivation (68% day 2, F3, 28=3.302, P=0.035), but no other significant differences were noted between the time points. Similarly, dystonin was lowered by 63% at day 14 time point, which was different compared to day 7 denervation time point (F3, 28=3.769, P=0.022). In contrast, MACF expression was upregulated fourfold after 3 days of denervation compared to all other time points (F3, 27=4.425, P=0.005).
Protein expression of plectin, dystonin, and MACF in denervated skeletal muscle
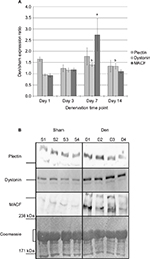
Protein levels were determined by “high molecular weight” Western blotting. Paired t-tests were performed to analyze the difference between plakin protein expression levels in den vs sham muscle at each time point. Overall, the protein levels for plectin remained similar between the sham and den samples over the denervation time course (Figure 3). The ANOVA, performed on the den/sham ratios, also revealed that the ratios were similar over the denervation time course (F3, 24=0.640, P=0.597). However, dystonin expression was unchanged after 1 day and 3 days of denervation (P=0.857 and P=0.077 respectively), a slight upregulation was detected after 7 days (50.2% increase, P=0.019) and 14 days of denervation (30.5% increase, P=0.005). Analyses of the den/sham ratios also indicate that the dystonin expression ratios were significantly greater at day 7 and day 14 postdenervation (F3, 28=5.083, P=0.006). As for MACF, protein expression was transiently upregulated in den muscles compared to sham at day 7 (67.4% increase, P=0.002), and the MACF den/sham expression ratios at day 7 were significantly greater than all other denervation time points (F3, 28=4.663, P=0.009) (Figure 3).
Discussion
The main objective of this study was to determine how the gene expression of muscle plakin family members (plectin, dystonin, and MACF) is impacted by neuromuscular inactivation. We used the denervation model, which is the most extreme model of inactivation, because it ensures that all aspects of communication between the motor neuron and skeletal muscle (ie, electrical activation and neurotrophic interaction) are interrupted.
Consistent with previous studies, we observed a 10% decrease in mouse gastrocnemius weight after 3 days of denervation and a ~40% decrease in muscle weight after 14 days of denervation.25,30,32 Also in line with previous studies, we detected significant upregulation of myogenin and downregulation of AChE mRNA expression in denervated skeletal muscle.26,27,33–36 Since these markers of denervation were aligned with scientific evidence in the literature, we proceeded to determine the expression profile of the selected skeletal muscle plakin family members.
The extent to which plectin, dystonin, and MACF expressions are affected in skeletal muscles that have undergone denervation-induced neuromuscular inactivation has not been extensively documented in the literature. We therefore conducted a series of qPCR and Western blot experiments to measure the mRNA and protein levels of these plakin family members in sham vs denervated muscles. Different trends were observed for each of these plakins. At the mRNA level, plectin was progressively downregulated over the denervation time course, leading to a 46% reduction in expression after 7 days and a 58% reduction after 14 days. However, we were unable to detect differences in plectin expression at the protein level at any of the denervation time points. In comparison, dystonin mRNA expression was gradually downregulated by up to 50% at 14 days postdenervation, whereas a slight increase in dystonin protein levels of ~50% and 30% were measured at 7 days and 14 days postdenervation, respectively. Finally, the change in MACF mRNA and protein was similar but slightly different in magnitude. In particular, both mRNA and protein expressions peaked at 7 days of denervation but mRNA showed a 2.5-fold increase, while protein expression was enhanced by 67%. We were unable to detect significant upregulation in MACF protein expression at any other time point, but mRNA showed a roughly twofold increase at day 3 and day 14.
Widespread changes in gene expression in various models of muscle atrophy including denervation have led to the identification of key proteins such as atrogin-1 and MuRF1 that mediate protein degradation during muscle wasting.37,38 In addition, mRNA expression studies in rats using RNA fingerprinting reveal that the levels of calpain-3, myosin heavy chain IIB, and muscle-specific enolase mRNA are downregulated, while TRAP-2 and glutamine synthetase are upregulated in skeletal muscles denervated for 5 days.39 Other studies report that the expression of myosin and other related contractile proteins, such as actin and tropomyosin, is downregulated in denervated muscle.40–44 Disturbances in calcium balance, related to reduced calcium-ATPase activity, are also noted in denervated muscle.45–47 Furthermore, immunofluorescence studies have shown desmin and tubulin protein levels to be upregulated, whereas the giant cytoskeletal protein titin is downregulated.28–30 In another study, microarray analyses were used to survey the expression of hundreds of genes in short-term denervated adult mouse.48 The reporting criterion for genes that were upregulated was set at a minimum of 2× change in mRNA gene expression in the denervated sample vs the control sample, whereas the inclusion criterion for downregulated genes was set at ≤0.5× (denervated/control). By applying similar criteria (ie, minimum twofold increase and 0.5-fold decrease) to the present study, our plectin, dystonin, and MACF mRNA expression data were considered to be significantly different in denervated compared to innervated muscle. Our qPCR results for MACF are in-line with the microarray analyses performed using RNA isolated from either short-term (ie, 1 week) and long-term (ie, 30–90 days) denervated skeletal muscles.48 In these latter two studies, a threefold increase in MACF expression was reported in denervated muscle compared to innervated muscle. However, changes in dystonin and plectin gene expressions were not reported by Magnusson et al48 or Batt et al,44 suggesting that differences in dystonin and plectin gene expressions may not have met the threshold criteria to be reported. It is also possible that the reductions in dystonin and plectin mRNA expressions within the first 2 weeks as reported here using qPCR are marginally transient in denervated muscles.
Using Western blot analysis, the levels of dystrophin and two dystrophin-associated proteins, β-dystroglycan and adhalin, have been shown to be upregulated after 2 weeks of denervation in rat skeletal muscles.31 Dystrophin was reportedly increased marginally (1.5×), and β-dystroglycan protein levels were 2.5× higher in denervated muscle vs control muscle, but due to the low abundance of adhalin in control muscle, the change in expression of adhalin could not be as accurately measured.31 Furthermore, utrophin, a dystrophin homolog expressed in skeletal muscle, has been shown to be upregulated both at the mRNA and protein levels in 14-day denervated rodent muscle.49 For MACF, we observed a similar trend, whereby both mRNA and protein expressions for MACF were elevated in denervated muscle. In contrast, our dystonin and plectin mRNA expression data (reduction after 1 week and 2 weeks of denervation) do not align with our protein expression data (marginal increase in dystonin, no change in plectin). The 30%–50% increase in dystonin protein expression postdenervation would constitute a marginal change in protein expression and the extent to which this alteration has any substantial physiological relevance remains an open question.
Our observation that plectin and dystonin protein levels remain largely stable postdenervation and that MACF protein expression increases may suggest that these cytoskeletal proteins help to preserve muscle fiber structural integrity following short-term denervation. Dystonin and plectin share similar functions, such as binding to mitochondria and localizing in part at the Z-line in muscle.7 Interestingly, the muscle isoform of dystonin (dystonin-b) binds to plectin through their respective actin-binding domains, and in plectin-null mice, dystonin protein is less abundant.50 Therefore, binding of dystonin to plectin could foster protein stability postdenervation, thereby contributing to their relatively stable expression at the protein level.
MACF is highly expressed in the nervous system, skin, and skeletal muscle of embryonic day 17.5 mice, as well as in the lung right before birth.6 However, MACF expression in muscle plummets shortly after birth, while it remains strong in the nervous system, skin, and lungs.6 This MACF expression pattern in skeletal muscle during prenatal (higher) vs postnatal (lower) development may suggest that MACF plays a more significant role in muscle cells during embryonic development and that once innervation is fully established, MACF is not as critically important. Interestingly, MACF subdomains have been shown to interact with rapsyn, which is an element of the postsynaptic compartment of neuromuscular junctions involved in clustering and stabilizing acetylcholine receptors (AChR).51 However, whether MACF plays a role in stabilizing AChR clusters in skeletal muscle fibers has not been ascertained. In denervated skeletal muscle fibers, the expression of AChR-ε subunit is suppressed, and that of the embryonic AChR-γ variant is transiently reexpressed in extrasynaptic regions, peaking at ~5 days postdenervation and returning to levels observed in innervated muscle after 20 days postdenervation.52–54 The fact that denervation appears to “reboot” the developmental expression profile of MACF, even transiently, may suggest that MACF potentially contributes to organizing elements of the postsynaptic region of neuromuscular junctions prior to innervation. Additional studies are required to confirm this notion.
In Batt et al,44 the authors sought to uncover a gene expression profile that “protects the reinnervation and functional recovery potential of denervated muscle”, in short-term (1 month) and long-term (3 months) denervated muscles. Their microarray analyses suggest that the expression of tubulin is maintained after 1 month of denervation. Our own qPCR data would also support that tubulin expression is maintained in short-term denervated muscle (Figure 1). In-line with this evidence, tubulin protein levels are reportedly maintained in fast muscles (plantaris and gastrocnemius) following 15 days of hindlimb unloading in rats.55 MACF is thought to be a key microtubule-interacting protein and is known to stabilize microtubule networks in a variety of cell types.56 It is, therefore, possible that the maintenance of tubulin expression and the microtubule network, within the initial weeks of denervation, is facilitated by interactions with MACF. Two additional studies report that tubulin protein accumulates or is more abundant in denervated, rodent fast-twitch muscles (ie, gastrocnemius muscle).29,57 This may be important to “maintain or preserve the integrity of the muscle fiber during progressive atrophy or regeneration”57 and “could be indicative of an adaptive mechanism, designed to maintain the integrity of the muscle fiber in view of eventual regenerative activities”.29 Both of these assertions support our hypothesis that cytoskeletal proteins (ie, possibly the interaction of MACF and microtubules) maintain some level of myofiber integrity during muscle atrophy.
Conclusion
Collectively, our results indicate that the mRNA and protein expressions of dystonin, plectin, and MACF are differentially regulated in skeletal muscles undergoing short-term denervation-induced atrophy. The stable protein expression levels of plectin and dystonin in combination with the enhanced levels of MACF protein during denervation, at a time when sarcomeric proteins are being lost, suggest that these plakin proteins are basic scaffolding elements of the muscle fiber that are necessary to facilitate recovery in the event of reinnervation.
Acknowledgments
The authors would like to thank Paul Michael for his technical assistance. The current article includes work based on PB’s Masters of Human Kinetics thesis submitted while he was a graduate student at Laurentian University.
Disclosure
The authors report no conflicts of interest in this work.
References
Bloch RJ, Gonzalez-Serratos H. Lateral force transmission across costameres in skeletal muscle. Exerc Sport Sci Rev. 2003;31(2):73–78. | ||
Bouameur JE, Favre B, Borradori L. Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases. J Invest Dermatol. 2014;134(4):885–894. | ||
Boyer JG, Bernstein MA, Boudreau-Larivière C. Plakins in striated muscle. Muscle Nerve. 2010;41(3):299–308. | ||
Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313(10):2189–2203. | ||
Reipert S, Steinböck F, Fischer I, Bittner RE, Zeöld A, Wiche G. Association of mitochondria with plectin and desmin intermediate filaments in striated muscle. Exp Cell Res. 1999;252(2):479–491. | ||
Bernier G, Pool M, Kilcup M, Alfoldi J, De Repentigny Y, Kothary R. Acf7 (MACF) is an actin and microtubule linker protein whose expression predominates in neural, muscle, and lung development. Dev Dyn. 2000;219(2):216–225. | ||
Dalpé G, Mathieu M, Comtois A, et al. Dystonin-deficient mice exhibit an intrinsic muscle weakness and an instability of skeletal muscle cytoarchitecture. Dev Biol. 1999;210(2):367–380. | ||
Ferrier A, Boyer JG, Kothary R. Cellular and molecular biology of neuronal dystonin. Int Rev Cell Mol Biol. 2013;300:85–120 | ||
Chen HJ, Lin CM, Lin CS, Perez-Olle R, Leung CL, Liem RK. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006;20(14):1933–1945. | ||
Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135(1):137–148. | ||
Goryunov D, He CZ, Lin CS, Leung CL, Liem RK. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol Cell Neurosci. 2010;44(1):1–14. | ||
Kothary R, Clapoff S, Brown A, Campbell R, Peterson A, Rossant J. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature. 1988;335(6189):435–437. | ||
Charlesworth A, Gagnoux-Palacios L, Bonduelle M, Ortonne JP, De Raeve L, Meneguzzi G. Identification of a lethal form of epidermolysis bullosa simplex associated with a homozygous genetic mutation in plectin. J Invest Dermatol. 2003;121(6):1344–1348. | ||
Fuchs P, Zörer M, Rezniczek GA, et al. Unusual 5’ transcript complexity of plectin isoforms: novel tissue-specific exons modulate actin binding activity. Hum Mol Genet. 1999;8(13):2461–2472. | ||
Rezniczek GA, Konieczny P, Nikolic B, et al. Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with β-dystroglycan. J Cell Biol. 2007;176(7):965–977. | ||
Winter L, Abrahamsberg C, Wiche G. Plectin isoform 1b mediates mitochondrion-intermediate filament network linkage and controls organelle shape. J Cell Biol. 2008;181(6):903–911. | ||
Powers SK, Wiggs MP, Duarte JA, Zergeroglu AM, Demirel HA. Mitochondrial signaling contributes to disuse muscle atrophy. Am J Physiol Endocrinol Metab. 2012;303(1):E31–E39. | ||
Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37(10):1974–1984. | ||
Kandarian SC, Jackman RW. Intracellular signaling during skeletal muscle atrophy. Muscle Nerve. 2006;33(2):155–165. | ||
Andrianjafiniony T, Dupré-Aucouturier S, Letexier D, Couchoux H, Desplanches D. Oxidative stress, apoptosis, and proteolysis in skeletal muscle repair after unloading. Am J Physiol Cell Physiol. 2010;299(2):C307–C315. | ||
Kelleher AR, Kimball SR, Dennis MD, Schilder RJ, Jefferson LS. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab. 2013;304:E229–E236. | ||
Adhihetty PJ, O’Leary MF, Chabi B, Wicks KL, Hood DA. Effect of denervation on mitochondrially mediated apoptosis in skeletal muscle. J Appl Physiol. 2007;102(3):1143–1151. | ||
Lawler JM, Song W, Demaree SR. Hindlimb unloading increases oxidative stress and disrupts antioxidant capacity in skeletal muscle. Free Radic Biol Med. 2003;35(1):9–16. | ||
Houston FE, Hain BA, Adams TJ, Houston KL, O’Keeffe R, Dodd SL. Heat shock protein 70 overexpression does not attenuate atrophy in Botulinum neurotoxin type A treated skeletal muscle. J Appl Physiol. 2015;119(1):83–92. | ||
Hyatt J-PK, Roy RR, Baldwin KM, Edgerton VR. Nerve activity-independent regulation of skeletal muscle atrophy: role of MyoD and myogenin in satellite cells and myonuclei. Am J Physiol Cell Physiol. 2003;285(5):C1161–C1173. | ||
Eftimie R, Brenner HR, Buonanno A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc Natl Acad Sci U S A. 1991;88(4):1349–1353. | ||
Adams L, Carlson BM, Henderson L, Goldman D. Adaptation of nicotinic acetylcholine receptor, myogenin, and MRF4 gene expression to long-term muscle denervation. J Cell Biol. 1995;131(5):1341–1349. | ||
Tews DS, Goebel HH, Schneider I, Gunkel A, Stennert E, Neiss WF. Expression profile of stress proteins, intermediate filaments, and adhesion molecules in experimentally denervated and reinnervated rat facial muscle. Exp Neurol. 1997;146(1):125–134. | ||
Boudriau S, Côté CH, Vincent M, Houle P, Tremblay RR, Rogers PA. Remodeling of the cytoskeletal lattice in denervated skeletal muscle. Muscle Nerve. 1996;19(11):1383–1390. | ||
Chen SP, Sheu JR, Lin AC, Hsiao G, Fong TH. Decline in titin content in rat skeletal muscle after denervation. Muscle and Nerve. 2005;32(6):798–807. | ||
Biral D, Senter L, Salviati G. Increased expression of dystrophin, beta-dystroglycan and adhalin in denervated rat muscles. J Muscle Res Cell Motil. 1996;17(5):523–532. | ||
Sacheck JM, Hyatt JP, Raffaello A, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21(1):140–155. | ||
Sketelj J, Crne-Finderle N, Strukelj B, Trontelj JV, Pette D. Acetylcholinesterase mRNA level and synaptic activity in rat muscles depend on nerve-induced pattern of muscle activation. J Neurosci. 1998;18(6):1944–1952. | ||
Pregelj P, Trinkaus M, Zupan D, Trontelj JJ, Sketelj J. The role of muscle activation pattern and calcineurin in acetylcholinesterase regulation in rat skeletal muscles. J Neurosci. 2007;27(5):1106–1113. | ||
Boudreau-Larivière C, Chan RY, Wu J, Jasmin BJ. Molecular mechanisms underlying the activity-linked alterations in acetylcholinesterase mRNAs in developing versus adult rat skeletal muscles. J Neurochem. 2000;74(6):2250–2258. | ||
Buonanno A, Edmondson DG, Hayes WP. Upstream sequences of the myogenin gene convey responsiveness to skeletal muscle denervation in transgenic mice. Nucleic Acids Res. 1993;21(24):5684–5693. | ||
Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. | ||
Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98(25):14440–14445. | ||
Tang H, Cheung WM, Ip FC, Ip NY. Identification and characterization of differentially expressed genes in denervated muscle. Mol Cell Neurosci. 2000;16(2):127–140. | ||
Jakubiec-Puka A, Ciechomska I, Morga J, Matusiak A. Contents of myosin heavy chains in denervated slow and fast rat leg muscles. Comp Biochem Physiol B Biochem Mol Biol. 1999;122(3):355–362. | ||
Jakubiec-Puka A. Changes in myosin and actin filaments in fast skeletal muscle after denervation and self-reinnervation. Comp Biochem Physiol Part A Physiol. 1992;102(1):93–98. | ||
Furuno K, Goodman MN, Goldberg AL. Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J Biol Chem. 1990;265(15):8550–8557. | ||
Horinouchi H, Kumamoto T, Kimura N, Ueyama H, Tsuda T. Myosin loss in denervated rat soleus muscle after dexamethasone treatment. Pathobiology. 2005;72(3):108–116. | ||
Batt J, Bain J, Goncalves J, et al. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006;20(1):115–117. | ||
Kaprielian Z, Bandman E, Fambrough DM. Expression of Ca2+-ATPase isoforms in denervated, regenerating, and dystrophic chicken skeletal muscle. Dev Biol. 1991;144(1):199–211. | ||
Lucas-Heron B, Loirat MJ, Ollivier B, Leoty C. Calcium-related abnormalities in fast and slow denervated skeletal muscle in rats. Comp Biochem Physiol Part A Physiol. 1986;84(4):601–606. | ||
Loirat MJ, Lucas-Heron B, Ollivier B, Leoty C. Calcium binding protein changes of sarcoplasmic reticulum from rat denervated skeletal muscle. Biosci Rep. 1988;8(4):369–378. | ||
Magnusson C, Svensson A, Christerson U, Tågerud S. Denervation-induced alterations in gene expression in mouse skeletal muscle. Eur J Neurosci. 2005;21(2):577–580. | ||
Jasmin BJ, Alameddine H, Lunde JA, et al. Expression of utrophin and its mRNA in denervated mdx mouse muscle. FEBS Lett. 1995;374(3):393–398. | ||
Steiner-Champliaud MF, Schneider Y, Favre B, et al. BPAG1 isoform-b: complex distribution pattern in striated and heart muscle and association with plectin and α-actinin. Exp Cell Res. 2010;316(3):297–313. | ||
Antolik C, Catino DH, O’Neill AM, Resneck WG, Ursitti JA, Bloch RJ. The actin binding domain of ACF7 binds directly to the tetratricopeptide repeat domains of rapsyn. Neuroscience. 2007;145(1):56–65. | ||
Gu Y, Hall ZW. Immunological evidence for a change in subunits of the acetylcholine receptor in developing and denervated rat muscle. Neuron. 1988;1(2):117–125. | ||
Yampolsky P, Pacifici PG, Witzemann V. Differential muscle-driven synaptic remodeling in the neuromuscular junction after denervation. Eur J Neurosci. 2010;31(4):646–658. | ||
Witzemann V, Chevessier F, Pacifici PG, Yampolsky P. The neuromuscular junction: selective remodeling of synaptic regulators at the nerve/muscle interface. Mech Dev. 2013;130(6–8):402–411. | ||
Jee H, Sakurai T, Lim J, Hatta H. Changes in α B-crystallin, tubulin, and MHC isoforms by hindlimb unloading show different expression patterns in various hindlimb muscles. J Exerc Nutrition Biochem. 2014;18(2):161–168. | ||
Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115(3):343–354. | ||
Nagao SI, Kumamoto T, Masuda T, Ueyama H, Toyoshima I, Tsuda T. Tau expression in denervated rat muscles. Muscle Nerve. 1999;22(1):61–70. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.