Back to Journals » International Journal of Nanomedicine » Volume 15
Effects of miR-26a on Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells by a Mesoporous Silica Nanoparticle - PEI - Peptide System
Authors Yan J, Lu X, Zhu X, Hu X, Wang L, Qian J, Zhang F, Liu M
Received 27 August 2019
Accepted for publication 28 December 2019
Published 23 January 2020 Volume 2020:15 Pages 497—511
DOI https://doi.org/10.2147/IJN.S228797
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Jia Yan, Xiaoli Lu, Xinchen Zhu, Xiaokun Hu, Lili Wang, Jun Qian, Feimin Zhang, Mei Liu
Jiangsu Key Laboratory of Oral Diseases, Department of Prosthodontics, Affiliated Hospital of Stomatology, Nanjing Medical University, Nanjing 210029, People’s Republic of China
Correspondence: Feimin Zhang; Mei Liu
Jiangsu Key Laboratory of Oral Diseases, Department of Prosthodontics, Affiliated Hospital of Stomatology, Nanjing Medical University, Nanjing 210029, People’s Republic of China
Tel +86 25 8503 1831
Fax +86 25 8651 6414
Email [email protected]; [email protected]
Introduction: RNA-based therapy for bone repair and regeneration is a highly safe and effective approach, which has been extensively investigated in recent years. However, the molecular stability of RNA agents still remains insufficient for clinical application. High porosity, tunable size, and ideal biodegradability and biosafety are a few of the characters of mesoporous silicon nanoparticles (MSNs) that render them a promising biomaterial carrier for RNA treatment.
Materials and Methods: In this study, a novel miR-26a delivery system was constructed based on MSNs. Next, we assessed the miRNA protection of the delivery vehicles. Then, rat bone marrow mesenchymal stem cells (rBMSCs) were incubated with the vectors, and the transfection efficiency, cellular uptake, and effects on cell viability and osteogenic differentiation were evaluated.
Results: The results demonstrated that the vectors protected miR-26a from degradation in vitro and delivered it into the cytoplasm. A relatively low concentration of the delivery systems significantly increased osteogenic differentiation of rBMSCs.
Conclusion: The vectors constructed in our study provide new methods and strategies for the delivery of microRNAs in bone tissue engineering.
Keywords: nanocarrier, microRNAs, osteoinduction, tissue engineering
Introduction
Annually, more than 20 million people worldwide are affected by bone defects caused due to trauma, infection, tumor, or congenital diseases.1 Therapies for bone regeneration pose a great clinical challenge.2 Autogenous graft is currently regarded as the “gold standard” treatment for bone loss because of its characteristic features such as excellent osteogenesis, osteoconduction and osteoinduction.3 Nonetheless, the volume of bone that is possible to be harvested from the iliac crest is restricted, and complications such as morbidity at the harvest site, local hematoma, and remodeling issues of the implanted bone are few of the significant concerns in clinical practices.4,5 Because of the abovementioned limitations of autogenous graft, there is an urgent need to devise novel clinical therapeutic approaches or methods and/or efficacious and efficient components with improved bone regeneration.6 Bone tissue engineering (BTE) has emerged as a promising bone regeneration methodology as it is capable of providing sufficient mechanical strength and promoting vascularization for their biocompatibility, biodegradability, and porosity.7,8 Despite these merits, BTE still possesses a relatively limited bone regeneration efficiency. Various strategies have been developed to improve the bone regeneration capabilities of BTE scaffolds.9,10 Apart from modifying the scaffold material, incorporation of diverse bone regeneration promoters including drugs, bioactive molecules, and siRNA are also attracting increasing attention.11–13
MicroRNAs (miRNAs) are a type of small non-coding single-stranded RNAs that play a significant role in the cellular activities of a living organism.14 They act as regulators of expression of post-transcriptional genes by either inhibiting the translating process or degrading mRNA molecules corresponding to the target gene.15 The functions of several miRNAs in the bone regeneration process have already been investigated.16–18 MiR-26a has been confirmed to regulate multiple pathways of osteogenic differentiation and promote bone restoration.19,20 However, in the context of miRNA therapy, negatively charged miRNAs are ineffective in penetrating the cell membranes that are also negatively charged. Moreover, in the absence of structural or chemical modifications, binding of miRNAs to intended targets in vivo tends to be ineffective while degrading rapidly.21 Virus-facilitated gene delivery was also considered an interesting possibility for efficient gene therapy; however, toxicities of the virus vector provoke certain concerns.22,23 Therefore, novel delivery vectors that can penetrate cell membrane effectively, protect miRNAs from degradation while generate negligible toxicity are urgently needed.
Mesoporous silica nanoparticles (MSNs), which have high porosity and specific surface area, tunable size, ideal biodegradability, and biosafety, were well accepted as a perfect drug delivery vehicle or system, therefore a promising vehicle for gene transfection.24–26 Following the development of MSM-41-type MSN nano-vehicles for drug delivery in 2001, various types of MSNs have been designed on the basis of customized payloads that aim to deliver, e.g., bioactive molecules, plasmids, siRNA and DNA, and so on.27–30 Advances have been made particularly in the application of MSNs as BTE scaffolds or at least a part of scaffolds in the recent years.31 Yao et al developed a novel MSNs incorporated-3D nanofibrous gelatin scaffold for dual-delivery of BMP-2 and deferoxamine to conserve their abilities of angiogenesis and osteogenesis.32 Zhao et al synthesized nanoparticle osteogenic delivery systems, by covalently grafting BMP-2 peptide on the surface of nanoparticles and loading dexamethasone into the channel of MSNs, which were efficient in inducing osteoblast differentiation and bone regeneration effectively in vivo.33 Here, we hypothesize vectors on the basis of MSNs that are capable of protecting loaded miR-26a and transporting it across the cell membrane barrier effectively in a cytotoxic-free manner, eventually promoting osteogenic differentiation of stem cells.
For authenticating this hypothesis, an MSN-based vector system was developed in which miR-26a was effectively loaded for the first time. Thereafter, various effects of the system, such as toxicity, transfection efficiency, miRNA delivery, and osteogenic differentiation on the rBMSCs were evaluated. We used rBMSCs because of their pluripotent nature with high self-renewal and multidirectional differentiation potential.34,35 The rBMSCs, with these merits apart from being easily available and multipliable are therefore perceived as ideal seeding cells for tissue engineering and target cells for cell or gene therapy.36,37 First, the protection effectiveness of the vectors to the miRNA were validated and the transfection doses and time were optimized. A 12 h treatment with 20 μg/mL MSN-miR-26a complex was found to deliver the target microRNA into rBMSCs effectively with insignificant cytotoxicity. Second, to detect the location of miR-26a transported into the transfected cells confocal microscopic examination was performed. Then alkaline phosphatase (ALP) activity and mineralization characterization was carried out to verify the occurrence of osteogenic differentiation of rBMSCs. On the 7th and 14th days after transfection, upregulation of multiple key growth factors associated with osteogenesis were found both at gene and protein levels, confirming the activation of osteogenic signal pathways. Altogether, the effectiveness of MSN-miR-26a complex in promoting stem cells osteogenic differentiation was demonstrated by these results; therefore, paved the way for its further application in bone regeneration medicine.
Materials and Methods
Mesoporous silica nanoparticles (~90 nm) were procured from Xi’an ruixi Biological Technology Co., Ltd (Xi’an, China). Riobo (Guangzhou, China) supplied micrON rno-miR-26a-5p mimic, micrON miRNA mimic NC, fluorescein amidite (FAM) labeled rno-miR-26a-5p mimic, and Bulge-LoopTM miRNA qRT-PCR Starter Kits. The KALA peptide (WEAKLAKALAKALAKHLAKALAKALKACEA) was designed by Sangon Biotech (Shanghai, China). Sigma-Aldrich (Germany) provided branched polyethylenimine (PEI, MW = 25,000Da), Guanidine HCL (MW = 95.53), and N-succinimidyl-3-(2-pyridyldithiol) propionate (SPDP). Chemicals and reagents such as Phosphate buffer saline 1X (PBS; Thermo Fisher Scientific, Waltham, MA, USA), serum-free and animal protein-free cell freezing medium (New Cell & Molecular Biotech Co., Ltd, Suzhou, China), TritonX-100 (Solarbio Life Sciences, Beijing, China), bicinchoninic acid protein assay kit (Leagene Biotechnology, Beijing, China), ALP assay kit (Jiancheng Bioengineering Institute, Nanjing, China), BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime Biotechnology, Shanghai, China), tetramethylethylenediamine (TEMED; Thermo Fisher Scientific, Waltham, MA, USA), NON-Fat Powdered Milk (BBI Life Sciences Corporation, Shanghai, China), PageRuler Prestained Protein Ladder (Thermo Fisher Scientific, Waltham, MA, USA) were utilized in this study. Cell Counting Kit-8 (CCK-8), SDS-PAGE protein loading buffer, 30% Acr-Bis (29:1), 10% SDS, 1M Tris-HCL (pH = 8.8), 1M Tris-HCL (pH = 6.8), Glycine (MW = 75.07), and 4% paraformaldehyde were procured from Biosharp (Hefei, China). Phenylmethanesulfonyl fluoride (PMSF), RIPA lysis buffer, ECL Plus Detection Kit, and WB Primary Antibody Dilution Buffer were obtained from EnoGene (Nanjing, China). Cyagen Biosciences (Guangzhou, China) provided OriCell Sprague-Dawley (SD) rBMSCs, SD Rat Mesenchymal Stem Cell Complete Medium, SD Rat Mesenchymal Stem Cell Osteogenic Differentiation Medium, OriCell Trypsin-EDTA Solution, and Alizarin Red S (ARS). Takara Minibest universal RNA Extraction Kit, PrimerScript RT Master Mix, and SYBR Premix EX Taq II were delivered by Takara Bio Inc. (Kusatsu, Japan).
Preparation of MSN_miRNA@PEI-KALA Delivery System
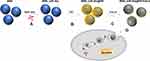
The researches of Li et al were referred to design MSN_miRNA@PEI-KALA.38,39 Specifically, 0.5 mg MSNs were added into a 1.5 mL microcentrifuge tube and dispersed in 80 μL ethanol, and 20 μL 4M Guanidine hydrochloride solution was added. To the tube were added 10 μL 0.1 nmol/μL miR-26a mimic or FAM-labeled miR-26a mimic or mimic NC or RNase-free H2O. The mixture was dispersed by an ultrasonic device (P = 80 W) for 10 min and was then shaken at a speed of 200 rpm at 4°C for 1 h. After being centrifuged at 12,000 rpm at 4°C for 10 min, the supernatant was removed and the sediment was rinsed once by ethanol. The sediment was resuspended in 100 μL ethanol, then well dispersed using the ultrasonic device for several minutes. Subsequently, an equal volume of 1 mg/mL PEI ethanol solution was added drop by drop. Following 15 min of ultrasonic dispersion, the mixture was centrifuged, supernatant segregated, and the sediment rinsed by ethanol. The thus attained particles were resuspended in 100 μL ethanol, followed by addition of 20 μL 0.2 mg/mL SPDP solution. This was followed by 15 min of ultrasonication and 30 min of precipitation; then, the mixture was centrifuged and the supernatant was discarded. Washed twice using RNase-free ddH2O, the particles were dispersed and 75 μL 1 mg/mL KALA peptide solution was added. Following dispersion by ultrasonication, the mixture was allowed to rest for 30 min and then centrifuged. It was rinsed twice using water, 500 μL ddH2O was added to the particles and the final concentration of the delivery system was measured to be 1 μg/μL. The vectors were stored at −20°C for later use. The flowchart illustrations of the preparation are shown in Figure 1.
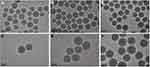
Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS)
The morphology and structure of MSNs were observed by TEM (JEM-2100; JEOL, Tokyo, Japan). The particles were dispersed in an appropriate solvent (e.g., deionized [DI] water or ethanol). Following ultrasonic dispersion for 15 min, the particle suspension was dripped onto a 200-mesh copper TEM grid (Electron Microscopy Sciences, Washington, PA) and dried at room temperature overnight. At least three images of each sample were gathered for statistical representation. Particle size and zeta potential in a liquid solution were measured using ZetaSizer Nano (Malvern Instruments Ltd., Worcestershire, UK).
Agarose Gel Electrophoresis Assay
Agarose gel electrophoresis was carried out to evaluate the miRNA protection of the deliveries. Before electrophoresis, we performed a series of treatments on the delivery systems. First, the particles were incubated with RNase A. Then, the particles were further treated with heparin and RNase A. Naked miRNA with or without RNase A treatment and the delivery systems with the foregoing treatments were loaded on a 1% agarose gel containing 0.01% GoldView with TAE running buffer at 100 V for 30 min. Consequently, the obtained bands were visualized by a UV (254 nm) illuminator and the images captured by Bio-Rad imaging system (Hercules, CA).
Cell Culture
SD rBMSCs were cultured at a temperature of 37°C in a cell incubator at a humidified atmosphere comprising 5% CO2. The rBMSCs were passaged on reaching ~80% confluence. Cells at passages 3–5 were used in this study.
Cell Viability Assay
SD rBMSCs were seeded in a 96-well plate with a concentration of 5 × 103 cells/well and incubated overnight prior to starting the treatments. The cells were washed with PBS and transferred to a low serum medium (5% FBS) without penicillin or streptomycin, prior to treating them with varying concentrations (5, 10, 20, 25, 50, and 100 μg/mL) of MSN@PEI-KALA, MSN_miR-26a@PEI-KALA and MSN_miR-NC@PEI-KALA. The CCK-8 assay was used to assess the cell viability at 12 h and 24 h post transfection. The cells without any treatment were set as control. Briefly, CCK-8 solution was mixed with the culture medium at a volume ratio of 1:10, and the thus obtained mixture was added to each well and incubated at 37°C for 2 h. Wells containing only CCK-8 solution and medium were set as blank. The absorbance of all samples was measured at 450 nm using a microplate reader (Spectra Max 190; Molecular Devices LLC, Sunnyvale, CA, USA). The cell viability was obtained in terms of percentages using the following formula:
Cell viability (%) = (OD test – OD blank)/(OD control – OD blank) × 100%
Confocal Laser Scanning Microscopy (CLSM) Observation
Glass-bottomed Petri dishes were used to seed rBMSCs and incubated overnight. The vehicles loaded with FAM-labeled miRNA molecules were added with a final concentration of 20 μg/mL. After allowing different incubation time periods, the medium was replaced with the prewarmed (37°C) Lyso-Tracker Red (Solarbio Life Sciences) probe-containing medium to label the lysosome. The loading solution was removed after an hour and the cells were washed twice with PBS. They were then fixed with 4% paraformaldehyde and washed twice with PBS; the cells were dyed with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime Biotechnology) to label the cell nuclei. Fluorescence images were captured using CLSM (Zeiss-LSM710; Carl Zeiss Meditec AG, Jena, Germany).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
To estimate the mRNA expression level of miR-26a, the rBMSCs were seeded at 1 × 105 cells/well in a 6-well plate, transferred to a low-serum DMEM on the second day and incubated with 20 μg/mL MSN_miR-26a@PEI-KALA, MSN@PEI-KALA, and MSN_miR-NC@PEI-KALA for different durations, and the cells without treatment were set as control.
To assess the mRNA expression level of osteogenic differentiation markers, including Runx2, Opn, Osx, and Bmp2 in various groups on the 7th and 14th days after osteogenic induction; the cells were seeded at 1 × 105 cells/well in a 6-well plate. Following the incubation with 20 μg/mL MSN_miR-26a@PEI-KALA, MSN_miR-NC@PEI-KALA, and MSN@PEI-KALA for 12 h each, the medium was changed to osteogenic differentiation medium. On the 7th and 14th days, the cells were collected and the total RNA isolated using the RNA extraction kit as per the manufacturer’s instructions. A total of 500 ng of RNA was reversely transcribed into cDNA using a reverse transcription kit, and qRT-PCR was carried out using the ABI 7900 Real-Time PCR System (Thermo Fisher Scientific). The primer sequences of osteogenic differentiation markers are provided in Table 1. The Bulge-LoopTM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for miR-26a were procured from RiboBio Biotech (Guangzhou, China); and the reference genes were GAPDH and U6. The relative expression level of the target gene was calculated according to equation 2^−ΔΔCT.
 |
Table 1 Primer Sequences for qRT-PCR Analysis |
ALP Staining and Activity Assays
The rBMSCs were seeded in a 24-well plate with 5 × 104 per well. MSN@PEI-KALA, MSN_miR-26a@PEI-KALA, and MSN_miR-NC@PEI-KALA were added in the well with a final concentration of 20 μg/mL. After 12 h the osteogenic induction medium was replaced. On the days 7 and 14 of osteogenic induction, the cells were dyed as per the manufacturer’s instructions in ALP staining kit. Briefly, the cells were first fixed with 4% paraformaldehyde and then rinsed twice with PBS. They were dyed using BCIP/NBT reagents at room temperature and washed with double distilled water for observation.
For the ALP activity assay, the rBMSCs were seeded in a 6-well plate and treated with vectors as described hereinbefore. After osteogenic induction for 7 and 14 days, the cells were lyzed using 0.5% TritonX-100. Post centrifugation, the supernatant was collected for subsequent experiments. The total protein concentration and ALP activity were detected following the instructions in the bicinchoninic acid protein assay and ALP assay kits, respectively. The ALP activity was normalized to the total protein content.
Matrix Mineralization Assay
Matrix mineralization level was evaluated using Alizarin red staining. The grouping and treatments of cells in this experiment were consistent with those used in the ALP staining. Fourteen days after the osteogenic induction, rBMSCs were fixed with 4% paraformaldehyde for 15 min and dyed with alizarin red solution for 5 min. They were then washed twice with double distilled water for observation. Then, 10% cetylpyridinium chloride (Bomei Biotechnology, Hefei, China) was added into the wells to dissolve the red matrix precipitate and sustained for 15 min. The optical density of the solution was read at 550 nm with a microplate reader.
Western Blot Analysis
To assess the protein levels of OPN, OSX, and BMP2 in rBMSCs treated with MSN@PEI-KALA, MSN_miR-26a@PEI-KALA, and MSN_NC@PEI-KALA Western blot was carried out. Proteins, extracted from each group using RIPA lysis buffer and 1% PMSF, were loaded onto 10% SDS-PAGE gel with equal amounts for electrophoresis. Proteins were then transferred onto a polyvinylidene difluoride membrane. After being blocked with 5% nonfat milk for 2 h, the membranes were incubated using anti-Osteopontin/OPN antibody (Novusbio; 1:1000 dilution), anti-Osterix antibody (Abcam; 1:1000 dilution), and anti-BMP2 antibody (Abcam; 1:1000 dilution) at 4°C overnight, followed with suitable secondary antibodies that conjugated with horseradish peroxidase for 50 min at room temperature. The internal control was GAPDH. After rinsing the blots thrice, enhanced chemiluminescence kit was used to visualize the proteins, and the results were analyzed using the Image J (NIH, Bethesda, USA).
Statistical Analysis
All experimental data were expressed in terms of mean ± standard deviation (SD) values, and analyzed with the help of SPSS v.19.0 software (IBM Corporation, Armonk, NY, USA). Comparisons between groups were made using one-way ANOVA followed by the least significant difference (LSD) test. Statistical significance was set at p < 0.05.
Results and Discussion
Characterization and miRNA Protection of MSN_miR-26a@PEI-KALA Delivery Vehicles
First, the size and surface morphology of MSNs at various stages of loading were observed by TEM. As shown in Figure 2, the size of both particles and pores were homogeneous when no modifications (Figure 2A and D). The size of the naked particles was ~90 nm. After coating with PEI, the oily substance could be observed (indicated by red arrows) on the surface of MSNs and the size of the particles increased to ~120 nm, which led to agglomeration and poor dispersion of particles (Figure 2B and E). The δ potential of the particle changed from negative to positive (Table 2), thus confirming the successful modification of PEI onto MSNs. The TEM image of MSN_miR-26a@PEI-KALA shown in Figure 2C and F demonstrated that PEI was almost invisible and the agglomeration of MSNs was remarkably mitigated, suggesting the KALA peptides were conjugated onto the surface of MSN_miR-26a@PEI. Compared with MSN_miR-26a@PEI, the size of MSN_miR-26a@PEI-KALA was reduced (~109 nm) and the positive potential was increased, providing basic conditions allowing the carrier to penetrate the cell membrane. The size and zeta potential of the three kinds of particles measured by DLS experiment are given in Table 2. The loading efficiency of miRNA by MSNs was determined by the consumption of miRNA in the solution before and after adsorption. In our work, the amount of miRNA loaded by MSNs was about 14.95 μg miRNA/mg MSNs, with equivalent ~ 1.44 nmol miRNA/mg MSNs.
 |
Table 2 Size and Zeta Potential of the Three Particles in Respective Solutions |
To verify whether the delivery vehicles could protect miRNA from degradation, agarose gel electrophoretic analysis was performed to characterize the release of miRNA on various conditions.38,40 As indicated in Figure 3, almost no miRNA release was observed when no treatment was performed on deliveries (Lane 3). Similarly, being treated with RNase A did not cause any release of miRNA (Lane 4) either. Contrarily, the brand appeared on the agarose gel when the vehicles were further treated with heparin that triggers the release of miRNA (Lane 5), and then disappeared when RNase A (Lane 6), which degraded the released miRNA, was added into the mixture. These results revealed the miRNA protection effects of MSN_miR-26a@PEI-KALA.
Transfection Efficiency and Cytotoxicity of MSN_miR-26a@PEI-KALA Delivery Vehicles
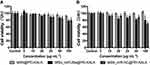
To determine the appropriate time and dosage of transfection, cell viability assay and qRT-PCR were performed. rBMSCs were incubated with varying concentrations of the particles for 12 h and 24 h, and then the cell viability was quantified by using the CCK-8 assay (Figure 4). The results exhibited that the cell survival rate decreased in a particle concentration dependent manner, and the cell viabilities at 24 h after incubation were generally lower than those observed at 12 h.
At 12 h, MSN_miR-26a@PEI-KALA and MSN_miR-NC@PEI-KALA displayed negligible cytotoxicity to rBMSCs at the dosage of 20 μg/mL compared with control. Nevertheless, even a relatively lower concentration (10 μg/mL) revealed significant cytotoxicity at 24 h. Furthermore, the survival rate of the cells co-cultured with MSN@PEI-KALA was not influenced either by the concentration or culture time. The results established that the vector itself had no cytotoxicity and the adverse effect on cell activity was mainly due to the transfection of miR-26a or miR-NC. Overall, the suitable time of incubation could be 12 h, and 20 μg/mL may be a safe concentration.
The results of FCM authenticated that the transfection efficiency increased as the dose (of delivery system) increased (Supplementary Figure 1A and B). The rBMSCs transfected with 20 μg/mL MSN_miR-26a@PEI-KALA exhibited a fluorescence intensity of ~23% at 12 h (Supplementary Figure 1C).
As displayed in Figure 5, after rBMSCs were incubated with 20 μg/mL MSN_miR-26a@PEI-KALA for varying time periods, the expression levels of miR-26a in each group was significantly increased compared with that in the control, and there was no significant difference between the control and the cells treated with MSN_miR-NC@PEI-KALA and MSN@PEI-KALA. The highest increment was found particularly in the 12 h group, indicating that maximal delivery efficiency was reached post 12 h transfection. These results are consistent with those obtained from the CCK-8 assay and FCM, suggesting that 12 h is the optimal transfection time, beyond which the transfection efficiency might be compromised by the cytotoxicity of MSNs vectors.
On the basis of the abovementioned experiments, the transfection concentration of 20 μg/mL and the incubation time of 12 h were opted for the following experiments.
Cellular Uptake and Location of miRNA
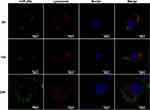
As presented, the protective effects of the miRNA from degradation in vitro by the vehicles and observed negligible side effect on cell activity with appropriate treatments, we then investigated the cellular uptake and the localization of miRNAs in the cell. The rBMSCs were incubated with FAM-labeled miRNA molecules encapsulated in deliveries at a concentration of 20 μg/mL, for varying incubation periods followed by CLSM observation. As shown in Figure 6, post incubation for 6 h, green fluorescence was observed in the cytoplasm, the distribution of which was granular and concentrated, and overlapped with the red fluorescence, suggesting that MSN_miR-26a@PEI-KALA had been internalized by cells and entered the lysosome. KALA is an amphipathic peptide with strong membrane activity.41 Several studies have evidenced that KALA promotes endosome formation and delivers the gene via the endosomal pathway.42 On the basis of the positive surface charge of the carriers and the unique properties of KALA peptides, it is inferred that the entrance of deliveries into cells may follow the pathway as follows;43 first, the opposite surface charge of vehicles (positive) and cell membrane (negative) drive the interaction between each other; next, the hydrophobic interaction between the leucine-rich side of KALA and the lipid side chain of the plasma membrane promotes the embedding of vehicles into the lipid bilayer; eventually, MSN_miR-26a@PEI-KALA are transported to the lysosome via the endosomal pathway.
Following incubation for 12 h, part of the green fluorescence that separated from the red one was more dispersed; whereas, the green fluorescence was diffused throughout the cytoplasm for 24 h. Moreover, neither the red nor the green fluorescence overlapped the blue fluorescence, signifying that the miRNA molecules were gradually released from the lysosome into the cytoplasm. Since 1980s, it had been reported that pH-responsive amphiphilic fusion peptides (such as KALA) promoted the escape of gene vectors from the endosome into the cytoplasm, owing to their membrane disrupting activity.44,45 Furthermore, PEI was a cationic polymer with excellent endosomolytic capability, which lead to osmotic swelling and disruption of endosome for proton buffering effects.30,46 Considering the endosome disruptive properties of PEI and the improvement of delivery efficiency, a protein or peptide which is similar to natural component should be designed that interacts with membranes in a natural way and is active under the milder conditions to facilitate safer and more efficient delivery and release of therapeutic agents in the furture.
ALP Activity and Mineralization Evaluation
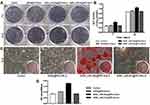
ALP was identified as a marker of early osteoblastic differentiation.47,48 ALP staining and activity assays were performed on days 7 and 14 after transfections to test the osteogenic differentiation level of rBMSCs. The highest ALP staining intensity was found in the group treated with MSN_miR-26a@PEI-KALA, whereas negligible difference was observed among the other three groups (Figure 7A). These results were in good agreement with those of the quantitative ALP activity assay and statistical significance was found (p < 0.05; Figure 7B). In addition, the level of ALP activity was observed to be higher on day 14 than on day 7. All these results indicated that the transfection with MSN_miR-26a@PEI-KALA promotes early osteoblastic differentiation of rBMSCs.
Alizarin red staining was performed to assess the degree of mineralization of the extracellular matrix after osteogenic induction for 14 days (Figure 7C). Calcium deposition of MSN_miR-26a@PEI-KALA treatment group was remarkably higher than those of other groups, consistent with the results of semi-quantification examination of ARS extracts (Figure 7D).
The results of ALP activity and mineralization evaluation confirmed that the delivery of MSN_miR-26a@PEI-KALA into rBMSCs indeed promoted the osteoblast differentiation both in early and later stages. It is conceivable that this was driven by miR-26a mimic in a manner consistent with that in earlier studies reporting miR-26a treatment could effectively improve the osteogenic differentiation of mesenchymal stem cells.49
PCR Analysis and Western Blot Analysis
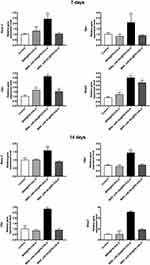
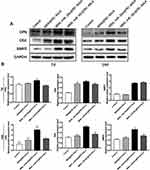
To further understand the mechanisms of how miR-26a-loaded MSNs promoted the osteogenic differentiation, several osteogenic gene expressions of rBMSCs on days 7 and 14 following osteogenic induction were tested. Studies have already revealed that miR-26a could promote the osteogenic differentiation potential of stem cells by regulating multiple osteogenic differentiation pathways and inducing the expression of several key growth factors related to osteogenesis.50 Runx2, specifically expressed in the initial stages of osteoblast differentiation that it was the earliest and most critical biomarker of bone formation or regeneration.51,52 Opn was the downstream target of Runx2 and regulated matrix mineralization.48,53 Osx, an essential transcription factor for osteoblastic differentiation, played a crucial role in the differentiation and maturation of osteoblasts.18 During the bone formation, BMP-2 or one of its co-signaling BMPs is imperative.54 In this case, the expression levels of these key osteogenic regulation genes were established and are shown in Figure 8. rBMSCs treated with MSN_miR-26a@PEI-KALA exhibited the highest levels of Runx2, Opn, Osx, and Bmp2 after osteogenic induction of both 7th and 14th days. There was no significant difference in the expression levels of Runx2 and Opn among the other three groups. Post osteogenic induction of 7 days, it was hypothesized that the higher expression level of the Osx among the three treatment groups than that in the control is associated with KALA peptides. Thus far, KALA peptides have not been shown to promote osteogenic differentiation by any study, which can be studied further in our future work. The cells treated with MSN_NC@PEI-KALA showed a significantly higher level of Bmp2, which is inferred may be related to some effects caused by miR-NC. The results of the Western blot were highly consistent with those of PCR (Figure 9).
The results of PCR and Western blot revealed that miR-26a mimics delivered by MSN_miR-26a@PEI-KALA could effectively promote the expression and secretion of multiple osteogenic regulators at gene as well as protein levels in rBMSCs.
Conclusion
It has been reported that miR-26a plays a critical role in regulating bone formation, and miR-26a could promote the osteogenic differentiation tendency of MSCs both in vitro and in vivo. However, further utilization of microRNAs in bone regeneration medicine is somewhat limited by its inadequate stability and high sensitivity for degradation in the physiological condition, and low cell membrane penetration capability, thus diminishing its effectiveness in the promotion of bone regeneration. Here, we designed a vector loaded with miR-26a mimics. MiR-26a was loaded into the channel of MSNs, and PEI was coated on the surface to protect miR-26a from degradation prior to entering into the target cells. It is proposed that the delivery system carrying miR-26a enters into the lysosome of the cells through endocytosis and escapes from the lysosome via the membrane fusion of KALA peptides and proton sponge mechanism of PEI, resulting in the release of miR-26a into the cytoplasm and eventually promoting the osteogenic differentiation of rBMSCs. As expected, even being treated by low concentrations of MSN_miR-26a@PEI-KALA with negligible cytotoxicity, considerable transfection efficiency can be achieved and therefore significant promotion of osteogenic differentiation of rBMSCs detected. These results authenticate the effectiveness of utilization of MSNs as a vector for the delivery of microRNAs to promote the osteogenic differentiation. For further investigations, it is hypothesized that the vectors can either be used alone or in combination with bone meal and periosteum, or the compound on the scaffold that is used in the bone defect region, to achieve sustained release of miRNA and long-term osteogenesis promotion. In addition, MSN_miR-26a@PEI-KALA has abundantly available groups on their surface, which can be further modified with functional polymers for active targeting of stem cells, enhanced environment responsiveness, and biocompatibility. Altogether, this work aimed to provide some novel insights in the application of microRNA for BTE.
Acknowledgment
This study was supported by grants from the National Key Research and Development Program of China (2016YFA0201704/2016YFA0201700), the Postdoctoral Foundation of Jiangsu (1701163B), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2014-37).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Habibovic P. (*) Strategic directions in osteoinduction and biomimetics. Tissue Eng Part A. 2017;23(23–24):1295–1296. doi:10.1089/ten.tea.2017.0430
2. Liu M, Lv Y. Reconstructing bone with natural bone graft: a review of in vivo studies in bone defect animal model. Nanomaterials (Basel). 2018;8(12):999. doi:10.3390/nano8120999
3. Martin V, Bettencourt A. Bone regeneration: biomaterials as local delivery systems with improved osteoinductive properties. Mat Sci Eng C-Mater. 2018;82:363–371. doi:10.1016/j.msec.2017.04.038
4. Ho-Shui-Ling A, Bolander J, Rustom LE, Johnson AW, Luyten FP, Picart C. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi:10.1016/j.biomaterials.2018.07.017
5. Sohn HS, Oh JK. Review of bone graft and bone substitutes with an emphasis on fracture surgeries. Biomater Res. 2019;23:9. doi:10.1186/s40824-019-0157-y
6. Iaquinta MR, Mazzoni E, Manfrini M, et al. Innovative biomaterials for bone regrowth. Int J Mol Sci. 2019;20(3):E618. doi:10.3390/ijms20030618
7. Majidinia M, Sadeghpour A, Yousefi B. The roles of signaling pathways in bone repair and regeneration. J Cell Physiol. 2018;233(4):2937–2948. doi:10.1002/jcp.26042
8. De Witte TM, Fratila-Apachitei LE, Zadpoor AA, Peppas NA. Bone tissue engineering via growth factor delivery: from scaffolds to complex matrices. Regen Biomater. 2018;5(4):197–211. doi:10.1093/rb/rby013
9. Sobacchi C, Erreni M, Strina D, Palagano E, Villa A, Menale C. 3D bone biomimetic scaffolds for basic and translational studies with mesenchymal stem cells. Int J Mol Sci. 2018;19(10):3150. doi:10.3390/ijms19103150
10. Chocholata P, Kulda V, Babuska V. Fabrication of scaffolds for bone-tissue regeneration. Materials (Basel). 2019;12(4):568. doi:10.3390/ma12040568
11. Kowalczewski CJ, Saul JM. Biomaterials for the delivery of growth factors and other therapeutic agents in tissue engineering approaches to bone regeneration. Front Pharmacol. 2018;9:513. doi:10.3389/fphar.2018.00513
12. Vieira S, Vial S, Reis RL, Oliveira JM. Nanoparticles for bone tissue engineering. Biotechnol Prog. 2017;33(3):590–611. doi:10.1002/btpr.2469
13. Zhang K, Jia Z, Yang B, et al. Adaptable hydrogels mediate cofactor-assisted activation of biomarker-responsive drug delivery via positive feedback for enhanced tissue regeneration. Adv Sci (Weinh). 2018;5(12):1800875. doi:10.1002/advs.201800875
14. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi:10.1016/S0092-8674(03)00231-9
15. Kittelmann S, McGregor AP. Modulation and evolution of animal development through microRNA regulation of gene expression. Genes (Basel). 2019;10(4):321. doi:10.3390/genes10040321
16. Lozano C, Duroux-Richard I, Firat H, Schordan E, Apparailly F. MicroRNAs: key regulators to understand osteoclast differentiation? Front Immunol. 2019;10:375. doi:10.3389/fimmu.2019.00375
17. Frohlich LF. MicroRNAs at the Interface between osteogenesis and angiogenesis as targets for bone regeneration. Cells. 2019;8(2):E121. doi:10.3390/cells8020121
18. Moghaddam T, Neshati Z. Role of microRNAs in osteogenesis of stem cells. J Cell Biochem. 2019;120(8):14136–14155. doi:10.1002/jcb.28689
19. Wang Z, Zhang D, Hu Z, et al. MicroRNA-26a-modified adipose-derived stem cells incorporated with a porous hydroxyapatite scaffold improve the repair of bone defects. Mol Med Rep. 2015;12(3):3345–3350. doi:10.3892/mmr.2015.3795
20. Zhang X, Li Y, Chen YE, Chen J, Ma PX. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat Commun. 2016;7:10376. doi:10.1038/ncomms10376
21. Tsekoura EK, Bahadur KCR, Uludag H. Biomaterials to facilitate delivery of RNA agents in bone regeneration and repair. ACS Biomater Sci Eng. 2017;3(7):1195–1206. doi:10.1021/acsbiomaterials.6b00387
22. Giacca M, Zacchigna S. Virus-mediated gene delivery for human gene therapy. J Control Release. 2012;161(2):377–388. doi:10.1016/j.jconrel.2012.04.008
23. Ajithkumar KC, Pramod K. Artificial virus as trump-card to resolve exigencies in targeted gene delivery. Mini-Rev Med Chem. 2018;18(3):276–286. doi:10.2174/1389557517666170529080316
24. Diab R, Canilho N, Pavel IA, Haffner FB, Girardon M, Pasc A. Silica-based systems for oral delivery of drugs, macromolecules and cells. Adv Colloid Interface Sci. 2017;249:346–362. doi:10.1016/j.cis.2017.04.005
25. Kesse S, Boakye-Yiadom KO, Ochete BO, et al. Mesoporous silica nanomaterials: versatile nanocarriers for cancer theranostics and drug and gene delivery. Pharmaceutics. 2019;11(2):77. doi:10.3390/pharmaceutics11020077
26. Rimpei K, Mitsuru N, Kanjiro M. Functionalization of silica nanoparticles for nucleic acid delivery. Nano Res. 2018;11(10):5219–5239. doi:10.1007/s12274-018-2116-7
27. Vallet-Regi M, Ramila A, Del Real RP, Perez-Pariente J. A new property of MCM-41: drug delivery system. Chem Mater. 2001;13(2):308–311. doi:10.1021/cm0011559
28. Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VS. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. J Am Chem Soc. 2004;126(41):13216–13217. doi:10.1021/ja046275m
29. Ashley CE, Carnes EC, Epler KE, et al. Delivery of small interfering RNA by peptide-targeted mesoporous silica nanoparticle-supported lipid bilayers. ACS Nano. 2012;6(3):2174–2188. doi:10.1021/nn204102q
30. Xia T, Kovochich M, Liong M, et al. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano. 2009;3(10):3273–3286. doi:10.1021/nn900918w
31. Shadjou N, Hasanzadeh M. Bone tissue engineering using silica-based mesoporous nanobiomaterials: recentprogress. Mater Sci Eng C Mater Biol Appl. 2015;55:401–409. doi:10.1016/j.msec.2015.05.027
32. Yao Q, Liu Y, Selvaratnam B, Koodali RT, Sun H. Mesoporous silicate nanoparticles/3D nanofibrous scaffold-mediated dual-drug delivery for bone tissue engineering. J Control Release. 2018;279:69–78. doi:10.1016/j.jconrel.2018.04.011
33. Zhou X, Feng W, Qiu K, et al. BMP-2 derived peptide and dexamethasone incorporated mesoporous silica nanoparticles for enhanced osteogenic differentiation of bone mesenchymal stem cells. ACS Appl Mater Interfaces. 2015;7(29):15777–15789. doi:10.1021/acsami.5b02636
34. Gardner OF, Alini M, Stoddart MJ. Mesenchymal stem cells derived from human bone marrow. Methods Mol Biol. 2015;1340:41–52. doi:10.1007/978-1-4939-2938-2_3
35. Andrzejewska A, Lukomska B, Janowski M. Concise review: mesenchymal stem cells: from roots to boost. Stem Cells. 2019;37(7):855–864. doi:10.1002/stem.3016
36. Ju C, Liu R, Zhang YW, et al. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother. 2019;115:108912. doi:10.1016/j.biopha.2019.108912
37. Perez JR, Kouroupis D, Li DJ, Best TM, Kaplan L, Correa D. Tissue engineering and cell-based therapies for fractures and bone defects. Front Bioeng Biotechnol. 2018;6:105. doi:10.3389/fbioe.2018.00105
38. Li X, Xie QR, Zhang J, Xia W, Gu H. The packaging of siRNA within the mesoporous structure of silica nanoparticles. Biomaterials. 2011;32(35):9546–9556. doi:10.1016/j.biomaterials.2011.08.068
39. Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. A mesoporous silica nanoparticle - PEI - Fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34(4):1391–1401. doi:10.1016/j.biomaterials.2012.10.072
40. Lee SH, Kim SH, Park TG. Intracellular siRNA delivery system using polyelectrolyte complex micelles prepared from VEGF siRNA-PEG conjugate and cationic fusogenic peptide. Biochem Biophys Res Commun. 2007;357(2):511–516. doi:10.1016/j.bbrc.2007.03.185
41. Wyman TB, Nicol F, Zelphati O, Scaria PV, Plank C, Szoka FC
42. Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151(3):220–228. doi:10.1016/j.jconrel.2010.11.004
43. Min SH, Lee DC, Lim MJ, et al. A composite gene delivery system consisting of polyethylenimine and an amphipathic peptide KALA. J Gene Med. 2006;8(12):1425–1434. doi:10.1002/jgm.973
44. Nouri FS, Wang X, Dorrani M, Karjoo Z, Hatefi A. A recombinant biopolymeric platform for reliable evaluation of the activity of pH-responsive amphiphile fusogenic peptides. Biomacromolecules. 2013;14(6):2033–2040. doi:10.1021/bm400380s
45. Mok H, Park TG. Self-crosslinked and reducible fusogenic peptides for intracellular delivery of siRNA. Biopolymers. 2008;89(10):881–888. doi:10.1002/bip.21032
46. Vermeulen LMP, De Smedt SC, Remaut K, Braeckmans K. The proton sponge hypothesis: fable or fact? Eur J Pharm Biopharm. 2018;129:184–190. doi:10.1016/j.ejpb.2018.05.034
47. Hu S, Chen H, Zhou X, et al. Thermally induced self-agglomeration 3D scaffolds with BMP-2-loaded core-shell fibers for enhanced osteogenic differentiation of rat adipose-derived stem cells. Int J Nanomedicine. 2018;13:4145–4155. doi:10.2147/IJN.S167035
48. Chen X, Wang L, Zhao K, Wang H. Osteocytogenesis: roles of physicochemical factors, collagen cleavage and exogenous molecules. Tissue Eng Part B Rev. 2018;24(3):215–225. doi:10.1089/ten.teb.2017.0378
49. Li Y, Fan L, Hu J, et al. MiR-26a rescues bone regeneration deficiency of mesenchymal stem cells derived from osteoporotic mice. Mol Ther. 2015;23(8):1349–1357. doi:10.1038/mt.2015.101
50. Li Y, Fan L, Liu S, et al. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34(21):5048–5058. doi:10.1016/j.biomaterials.2013.03.052
51. Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Gene Dev. 2001;15(4):467–481. doi:10.1101/gad.845101
52. Zhang X, Aubin JE, Inman RD. Molecular and cellular biology of new bone formation: insights into the ankylosis of ankylosing spondylitis. Curr Opin Rheumatol. 2003;15(4):387–393. doi:10.1097/00002281-200307000-00004
53. Martin V, Ribeiro IA, Alves MM, et al. Engineering a multifunctional 3D-printed PLA-collagen-minocycline-nanoHydroxyapatite scaffold with combined antimicrobial and osteogenic effects for bone regeneration. Mater Sci Eng C Mater Biol Appl. 2019;101:15–26. doi:10.1016/j.msec.2019.03.056
54. Blair HC, Larrouture QC, Li Y, et al. Osteoblast differentiation and bone matrix formation in vivo and in vitro. Tissue Eng Part B Rev. 2017;23(3):268–280. doi:10.1089/ten.teb.2016.0454
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.