Back to Journals » Drug Design, Development and Therapy » Volume 9
DNAzyme-based probe for circulating microRNA detection in peripheral blood
Authors Shao G, Ji S, Wu A, Liu C, Wang M, Zhang P, Jiao Q, Kang Y
Received 29 May 2015
Accepted for publication 29 July 2015
Published 16 November 2015 Volume 2015:9 Pages 6109—6117
DOI https://doi.org/10.2147/DDDT.S89560
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Wei Duan
Guoli Shao,1,* Shufeng Ji,1,* Aiguo Wu,1 Cuiping Liu,2 Mengchuan Wang,1 Pusheng Zhang,1 Qingli Jiao,1 Yuzhan Kang2
1Department of General Surgery, 2Department of Critical Care Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Background: The recent discovery of microRNAs (miRNAs) and their extracellular presence suggest a potential role of these regulatory molecules in defining the metastatic potential of cancer cells and mediating the cancer–host communication. This study aims to improve the sensitivity of miRNA detection via DNAzyme-based method and enhance the selectivity by using the DNAzyme-based probe to reduce nonspecific amplification.
Methods: The miRNA probes were chemically synthesized with a phosphate at the 5' end and purified by polyacrylamide gel electrophoresis. Exosomal RNA from peripheral blood was isolated. Carboxylated magnetic microsphere beads (MBs) were functionalized with streptavidin (SA) according to a previously reported method with some modification. T capture probe-coated SA-MBs (DNA-MBs) were also prepared. The fluorescent spectra were measured using a spectrofluorophotometer.
Results: We designed an incomplete DNAzyme probe with two stems and one bubble structure as a recognition element for the specific detection of miRNA with high sensitivity. The background effects were decreased with increase of the added of DNA-MBs and capturing times. Therefore, 20 minutes was selected as the optimal concentration in the current study. The fluorescence intensity increases as the hybridization time changed and reached a constant level at 40 minutes, and 1 µM is the optimum signal probe concentration for self-assembled DNA concatemers formation. In the presence of miRNA, the fluorescence of the solution increased with increasing miRNA concentration. There is no obvious fluorescence in the presence of 10 mM of other nontarget DNA.
Conclusion: A simple, rapid method with high performance has been constructed based on identified circulating miRNA signatures using miRNA-induced DNAzyme. This assay is simple, inexpensive, and sensitive, enabling quantitative detection of as low as 10 fM miRNA.
Keywords: DNAzyme, miRNAs, breast cancer, detection
Introduction
Metastasis is the leading cause of mortality in breast cancer patients. Only 20% of the patients face a 5-year survival rate. Therefore, there is a great and urgent need to develop a predictive or early diagnostic method for the detection of metastasis that would allow the development of efficient treatment options.1
The recent discovery of microRNAs (miRNAs) and their extracellular presence suggest a potential role of these regulatory molecules in defining the metastatic potential of cancer cells and mediating the cancer–host communication.2 miRNAs are small noncoding RNAs that base-pair with the 3′ end untranslated regions (3′ UTRs) of protein-encoding mRNAs, resulting in mRNA destabilization and/or translational inhibition. The biogenesis of miRNAs is tightly controlled, and dysregulation of miRNAs is linked to cancer. miRNAs are also present extracellularly, either through binding to protein or lipid carriers or as a major RNA component of exosomes.
Recently, a few researches show that miR-105, which is characteristically expressed and secreted by metastatic breast cancer cells, can be detected in the circulation of the premetastatic stage. The levels of miR-105 in the blood are associated with ZO-1 expression and metastatic progression in early stage of breast cancer.3,4 In breast cancer patients, increased levels of miR-105 in the circulation can be detected at the premetastatic stage and correlate with the occurrence of metastasis, which strongly suggests that the clinical applications of miR-105 could act as a predictive or early diagnostic blood-borne marker as well as a therapeutic target for breast cancer metastasis.5,6
Previous studies by other groups have identified circulating miRNAs in the blood of cancer patients and their levels distinguish cancer patients from healthy controls.7–10 The aim of this study is to develop a predictive or early diagnostic method for the detection of cancer-secreted miRNAs. In this study, we aim to improve the sensitivity of miRNA detection via DNAzyme-based method and enhance the selectivity by using the DNAzyme-based probe to reduce nonspecific amplification.
Materials and methods
Oligonucleotides and RNA isolation
Oligonucleotides were synthesized by Thermo Fisher Scientific (Waltham, MA, USA). The sequences for the miR-105 detection are shown in Table 1. These probes were chemically synthesized with a phosphate at the 5′ end and purified by polyacrylamide gel electrophoresis (Thermo Fisher Scientific). Exosomal RNA from peripheral blood of 20 patients and ten healthy people was isolated according to the instruction of isolation kit.4 The ratio of the signal probe 1 and probe 2 has also been analyzed to obtain the optimized ratio. The results indicated that the optimized ratio of probe 1 compared to probe 2 is 1:1 (Figure S1).
  | Table 1 Sequences for the miRNA detection |
In this study, the sample collections have been approved by the ethics committee of Southern Medical University, Guangzhou, Guangdong, People’s Republic of China.
Preparation of streptavidin-coated magnetic microsphere beads
Carboxylated magnetic microsphere beads (MBs) (25 mL [1% w/v]) were functionalized with streptavidin (SA) (25 mL, 1 mg/mL), according to a previously reported method with some modification.4,11,12 Briefly, 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (20 mg/mL, 20 mL) and N-Hydroxysuccinimide (NHS) (10 mg/mL, 20 mL) were mixed with precooled 4-Morpholine Ethane Sulfonic Acid 9 (MES) buffer (25 mM, pH 5, 10 mL) and then slowly dripped into MBs solution and shaken gently for 30 minutes at room temperature to activate the carboxyl groups. After washing three times with MES buffer (25 mM, pH 5, 10 mL), SA was slowly added and shaken gently for another 30 minutes. Finally, the SA-coated MBs (SA-MBs) were blocked with bovine serum albumin ([BSA] buffer (50 mM Tris-HCl, 0.1% Tween-20, 0.5% BSA, pH 7.4) for 30 minutes. After magnetic collection, the SA-MBs were resuspended in Tris-BSA buffer and stored at 4°C before use.
Preparation of capture probe-coated SA-MBs
Biotinylated capture DNA was added into SA-MBs solution with a final concentration of 1 mM, and gently shaken for 30 minutes at room temperature.5,6 The captured DNA functionalized MBs were magnetically washed three times with BSA buffer (50 mM Tris–HCl, 0.1% Tween-20, 0.5% BSA, pH 7.4) and magnetically collected, the DNA-MBs were resuspended in Tris-BSA buffer and stored at 4°C before usage.
T4 ligased and DNAzyme-based reactions
For miRNA detection, 2 μL miR-105 was mixed with 3 μL 10× reaction buffer (330 mM Tris-acetate, 100 mM magnesium acetate [Mg(Ac)2], 660 mM potassium acetate [KAc], 1% Tween-20, and 10 mM dithiothreitol [DTT] [pH 7.9], 2 μL [10 U/μL] T4 ligase [Fermentas], 0.5 μL 10 μM deoxynucleotide triphosphates [dNTPs] mixture [Takara, Dalian, People’s Republic of China], and RNase-free water), 25 μL double distilled water. The reaction mixture was incubated at 37° for 30 minutes.
Measurement of fluorescent spectra
The DNAzyme-digested product was mixed with 10 μL 10× SYBR Green I (SG) dye (Thermo Fisher Scientific) and diluted to a final volume of 200 μL with 10 mM phosphate-buffered saline (PBS) (pH 7.4). The fluorescent spectra were measured using a spectrofluorophotometer. The excitation wavelength was 497 nm, and the spectra were recorded between 507 nm and 650 nm. The fluorescence emission intensity was measured at 530 nm.
Statistical analysis
Data were expressed as mean ± standard error of the mean. Statistical comparisons were performed using Student’s t-test. P-values <0.05 were considered statistically significant.
Results and discussion
Mechanism of DNAzyme-based probe for circulating microRNA detection
The schematic diagram of our method was shown in Figure 1; we designed an incomplete DNAzyme probe with two stems and one bubble structure as a recognition element for the specific detection of miR-105 with high sensitivity. The DNAzyme-based probe contains three domains: a miRNA-binding domain (the bubble structure), a DNAzyme domain, and a substrate domain. The specific probe–target binding occurs at the bubble structure, which contains one base miss that was specially designed to be complementary to the target miRNA. The target miR-105 binding leads to the formation of intact DNAzyme in the presence of T4 DNA ligase. In the presence of Cu2+ and ascorbate, the substrate is irreversibly cleaved at the cleavage site.12 The cleaved piece (in green) is released due to decreased affinity to the enzyme. The cleaved product is then applied to the formation of self-assembled DNA concatemers,12,13 eventually resulting amplified fluorescent signals in the presence of SYBR Green I.
Optimization of the experimental conditions
In order to solve the problem of false-positive result by the excess free substrate, which could hybridize with the capture DNA–MBs and construct the self-assembled DNA concatemers with signal probe 1 and signal probe 2. The captured DNA–MBs were used to first hybridize with the free substrate. Excess DNA–MBs were used for the capture of free substrate at different times. As shown in Figure 2, the background fluorescence was decreased with the increase of the added of DNA-MBs and capturing times. When the capturing time reached 20 minutes or higher, the fluorescence intensity became the lowest. Therefore, 20 minutes was selected as the optimal concentration in the current study. Also, the optimized concentration of signal probe 1 and signal probe 2 or hybridization time was optimized according to a previously reported method (Figure 3).14 As shown in Figure 3, the fluorescence intensity increased as the hybridization time changed and reached a constant level at 40 minutes, and the optimized hybridization time was shown at 45 minutes. Also, 1 μM is the optimum signal probe concentration for self-assembled DNA concatemers formation. We also investigated that the optimized hybridization time was shown at 37°C (data not shown).
Furthermore, we have also investigated the effects of Cu2+ and ascorbate on the FL intensity, respectively. The results indicated that the there were no significant differences among the different concentration of Cu2+ and ascorbate (Figure S2) at different capturing time. The result also showed that there were no significant differences among the different concentrations of Cu2+ and ascorbate in different ratio of probe 1 and probe 2 (Figure S3A) and at different hybridization time (Figure S3B). In the following study, we would investigate the other markers than Cu2+ and ascorbate in different conditions. We believe that the other markers would also be useful to the hybridization investigation.
Detection of miRNA under optimal experimental conditions
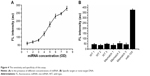
The sensitivity of this assay was done. Different concentrations of miR-105 (10 mM, 20 mM, 100 nM, 20 nM, 100 pM, 20 pM, 100 fM, 20 fM) were examined simultaneously. As shown in Figure 4A, in the absence of miR-105, the solution showed a very weak fluorescence. In the presence of miRNA, the fluorescence of the solution increased with increasing miR-105 concentration (Figure 4A). The limit of detection was 10 fM, which is calculated based on triple standard deviation from the mean of blanks.
Compared with other reported methods by other groups, the developed method exhibited a relatively high sensitivity for the detection of miR-105.15–17 Additionally, the present strategy possessed excellent reproducibility and none invasive to the patient. Moreover, these methods are time-saving and no complicated equipment is needed.
Moreover, our findings also proved that the different Cu2+ and ascorbate concentrations have not affected the fluorescence intensity in different miR-105 concentrations (Figure S4).
Specificity of the assay
To validate the specificity of the assay for miR-105 detection, several other nontarget DNA were tested, including wild-type DNA, DNA with one mismatch/two mismatches/three mismatches. As shown in Figure 4B, there is no obvious fluorescence in the presence of 10 mM of other nontarget DNA. These results indicated that our strategy exhibits high specificity for the detection of miRNA. In the following studies, we would attempt to explore the successful mechanism of dissociation, which would be good to confirm dissociation of the substrate strand after ascorbate addition.
Though the present study illustrated an effective probe for circulating miRNA detection of breast cancer in peripheral blood, there were also a few limitations. First, although the sensitivity and specificity for miR-105 were higher, the sensitivity and specificity for other miRNAs have not been explored. Second, as a promising applied method in clinics, the present method falls short in comparison with other previously applied methods.
In conclusion, we have successfully constructed a probe for circulating miRNA detection of breast cancer in peripheral blood. The reaction mechanism is based on the specific recognition of target DNA to DNAzyme and cleavage of DNAzyme. The released substrate triggers the formation of dsDNA concatamers by using SYBR Green I as a signal reporter. In comparison with the previously reported method, this method has the following advantages: 1) high sensitivity to detect a minimum of 10 fM miRNA,18 2) high selectivity,19 and 3) no requirement for complex and expensive instrumentation (except for a fluorescence spectrophotometer). The proposed method can be further used to screen the anticancer drugs and might provide a promising approach for the discovery of new anticancer drugs.
Acknowledgment
This work was financially supported by the Guangdong Province Science and Technology Plan Project (Grant no 00131170223726052).
Disclosure
The authors report no conflicts of interest in this work.
References
Mao J, Ladd J, Gad E, et al. Mining the pre-diagnostic antibody repertoire of TgMMTV-neu mice to identify autoantibodies useful for the early detection of human breast cancer. J Transel Med. 2014;12:121. | ||
Ohyashiki JH, Ohtsuki K, Mizoguchi I, et al. Downregulated microRNA-148b in circulating PBMCs in chronic myeloid leukemia patients with undetectable imnimal disease: a possible biomarker to discontinue imatinib safely. Drug Des Devel Ther. 2014;8:1151–1159. | ||
Lokody I. Cancer genetics: exosomally derived miR-105 destroys tight junctions. Nat Rev Genet. 2014;15:362. | ||
Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroy vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. | ||
Lokody I. Genetics: exosomally derived miR-105 destroys tight junctions. Nat Rev Cancer. 2014;14:386–387. | ||
Rask L, Balslev E, Jorgensen S, et al. High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer. APMIS. 2011;119:663–673. | ||
Backes C, Leidinger P, Keller A, et al. Blood born miRNAs signatures that can serve as disease specific biomarkers are not significantly affected by overall fitness and exercise. PLoS One. 2014;9:e102183. | ||
Gall TM, Frampton AE, Krell J, Castellano L, Stebbing J, Jiao LR. Blood-based miRNAs as noninvasive diagnostic and surrogative biomarkers in colorectal cancer. Expert Rev Mol Diagn. 2013;13:141–145. | ||
Park IH, Kang H, Lee KS, Nam S, Ro J, Kim JH. Identification and clinical implications of circulating microRNA for estrogen receptor-positive breast cancer. Tumor Biol. 2014;35:12173–12180. | ||
Duroux-Richard I, Pers YM, Fabre S, et al. Circulating miRNA-125b is a potential biomarker predicting response to rituximab in rheumatoid arthritis. Mediators Inflamm. 2014;2014:342524. | ||
Lucas LJ, Han JH, Yoon JY. Using highly carboxylated micropheres to simplify immunoassays and enhance diffusional mixing in a microfluidic device. Colloids Surf B Biointerfaces. 2006;49:106–111. | ||
Wu W, Chen J, Fang Z, et al. A self-assembled deoxyribonucleic acid concatemer for sensitive detection of single nucleotide polymorphism. Anal Chim Acta. 2013;804:235–239. | ||
Jiang L, Peng J, Yuan R, et al. An aptasensing platform for simultaneous detection of multiple analytes based on the amplification of exonuclease-catalyzed target recycling and DNA concatemers. Analyst. 2013;138:4818–4822. | ||
Ge C, Chen J, Wu W, et al. An enzyme-free and label-free assay for copper (II) ion detection based on self-assembled DNA concatamers and Sybr Green I. Analyst. 2013;138:4737–4740. | ||
Zhang X, Wu D, Liu Z, et al. An ultrasensitive label-free electrochemical biosensor for microRNA-21 detection based on a 2′-O-methyl modified DNAzyme and duplex-specific nuclease assisted target recycling. Chem Commun (Camb). 2014;50:12375–12377. | ||
Gao P, Wei JM, Li PY, et al. Screening of deoxyribozyme with high reversal efficiency against multidrug resistance in breast carcinoma cells. J Cell Mol Med. 2011;15:2130–2138. | ||
Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15:5–17. | ||
Wang XP, Yin BC, Wang P, Ye BC. Highly sensitive detection of microRNAs based on isothermal exponential amplification-assisted generation of catalytic G-guadruplex DNAzyme. Biosens Bioelectron. 2013;42:131–135. | ||
Meng X, Zhou Y, Liang Q, et al. Electrochemical determination of microRNA-21 based on bio bar code and hemin/G-quadruplet DNAenzyme. Analyst. 2013;138:3409–3415. |
Supplementary materials
  | Figure S1 Observation of the optimized ratio of probe 1 and probe 2. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.