Back to Journals » Drug Design, Development and Therapy » Volume 10
Distinct role of the Fas rs1800682 and FasL rs763110 polymorphisms in determining the risk of breast cancer among Han Chinese women
Authors Wang M, Wang Z, Wang X, Jin T, Dai Z , Kang H, Guan H, Ma X, Liu X, Dai Z
Received 21 April 2016
Accepted for publication 25 May 2016
Published 25 July 2016 Volume 2016:10 Pages 2359—2367
DOI https://doi.org/10.2147/DDDT.S111084
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Wei Duan
Meng Wang,1,* Zheng Wang,2,* Xi-Jing Wang,1 Tian-Bo Jin,3 Zhi-Ming Dai,4 Hua-Feng Kang,1 Hai-Tao Guan,1 Xiao-Bin Ma,1 Xing-Han Liu,1 Zhi-Jun Dai1
1Department of Oncology, Second Affiliated Hospital of Xi’an Jiaotong University, 2Department of Medical Oncology, Xi’an Central Hospital, 3National Engineering Research Center for Miniaturized Detection Systems, School of Life Sciences, Northwest University, 4Department of Anesthesia, Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, People’s Republic of China
*These authors contributed equally to this work
Background: In recent years, studies have demonstrated that polymorphisms in the promoters of Fas and FasL are significantly associated with breast cancer risk. However, the results of these studies were inconsistent. This case–control study was performed to explore the associations between Fas rs1800682 and FasL rs763110 polymorphisms and breast cancer.
Materials and methods: A hospital-based case–control study of 560 Han Chinese females with breast cancer (583 controls) was conducted. The MassARRAY system was used to search for a possible association between the disease risk and the two single nucleotide polymorphisms, Fas rs1800682 and FasL rs763110. Statistical analyses were performed using SNPStats software to conduct Pearson’s chi-square tests in five different genetic models. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated after adjustment to age and body mass index. PHASE v2.1 software was used to reconstruct all common haplotypes.
Results: A statistically significant association was found between Fas rs1800682 and increased breast cancer risk (AG vs AA: OR =1.37, 95% CI =1.06–1.78; AA+AG vs GG: OR =1.32, 95% CI =1.04–1.66), and also it was found that the FasL rs763110 polymorphism may decrease the risk. Stratified analyses demonstrated that the rs763110 polymorphism was associated with lower breast cancer risk among postmenopausal females (heterozygote model: OR =0.69, 95% CI =0.49–0.97; dominant model: OR =0.70, 95% CI =0.51–0.96). The T allele of rs763110 was also associated with a decreased risk of lymph node metastasis (allele model: OR =0.75, 95% CI =0.57–0.97) and an increased risk of the breast cancer being human epidermal growth factor receptor 2 positive (allele model: OR =1.37, 95% CI =1.03–1.18). Moreover, haplotype analysis showed that Ars1800682Trs763110 was associated to a statistically significant degree with lower risk of breast cancer (OR =0.70, 95% CI =0.53–0.91).
Conclusion: These data suggest that the presence of Fas rs1800683 is an important risk factor for breast cancer, whereas FasL rs763110 may exert a protective effect against the onset of breast cancer.
Keywords: Fas, FasL, single nucleotide polymorphism, breast cancer, risk
Introduction
Despite the existence of a comprehensive management strategy against breast cancer, both the incidence and the mortality rates of the disease are increasing. In the United States alone, 246,660 new breast cancer cases are expected in 2016, which account for 29% of all estimated new cancer diagnoses in females.1 Although the morbidity rate of breast cancer is lower in People’s Republic of China compared with that in developed countries, since the 1990s, the incidence rate has been rising more than twice as fast as the worldwide average, resulting in its becoming the most common cancer in Chinese females.2 As is also the case for other malignancies, the development and progression of breast cancer comprise a complex process where genetic predisposition, environment, and lifestyle all play a role.3 For the past few years, the study of single nucleotide polymorphisms (SNPs) has been a focus area of oncological research into genetic factors, with respect to susceptibility, diagnosis, treatment, and prognosis of malignant tumors. Recent genome-wide association and large-scale replication studies have identified common variants in more than 70 loci associated with breast cancer.4
Apoptosis, or programmed cell death, is a vital physiological process subjected to gene regulation. Aberrant regulation of apoptosis contributes to the development and progression, but also the treatment, of multiple diseases, including cancer.5,6 Fas, a cell surface protein also known as TNFSF6/CD95/APO-1, and Fas Ligand (FasL), a type II membrane protein, belong to the tumor necrosis factor receptor superfamily7,8 and play an important role in regulating apoptosis, as their crosslinking initiates the signal cascade of programmed cell death.7 Therefore, mutation in the Fas and FasL genes may affect the apoptotic procedure and, thus, the development and progression of tumors. Several pharmacogenetic studies revealed the association of Fas/FasL SNPs with the response of cancer chemotherapy, showing mutation of Fas/FasL may be an indicator of cancer treatment.9,10
A large number of studies revealed that Fas/FasL gene polymorphisms are associated with susceptibility to various types of cancer, including cervical,11–14 pharyngeal,15–17 digestive,18–22 and breast cancer.23–28 Further studies demonstrated that circulating, soluble Fas (sFas) can inhibit Fas-mediated apoptosis by neutralizing the FasL or the anti-Fas antibodies,29 while increased levels of sFas have been observed in serum from patients with breast cancer.30 The most extensively investigated Fas/FasL polymorphisms are rs1800682 (−670A>G) in the promoter region of Fas and rs763110 (−844C>T) in the promoter region of FasL. However, the association of Fas and FasL gene polymorphisms with breast cancer has not been irrefutably established as different studies often produce conflicting results. For example, the study performed by Xu et al28 showed that the Fas rs1800682 and FasL rs763110 polymorphisms may reduce the risk of breast cancer, whereas Hashemi et al27 reported that the same SNPs were significantly associated with an increased risk of breast cancer (odds ratio, OR =3.18, P=0.019; OR =2.40, P=0.024, respectively), while Crew et al23 found no significant association between these two genetic polymorphisms and breast cancer risk. Thus, this case–control study was conducted to explore the role of these two polymorphisms (Fas rs1800682 and FasL rs763110) in breast cancer risk, in a Chinese population.
Materials and methods
Study population
Patients who had breast cancer and were being treated at the Department of Oncology, the Second Affiliated Hospital, Xi’an Jiaotong University, were enrolled from January 2013 to October 2014. All the cases were verified using pathology and detailed immunohistochemical analysis, as described in our previous studies.31–33 Patients with prior cancers or lacking a detailed personal and clinical background were excluded. Ultimately, 560 breast cancer cases were enrolled in this study; 583 healthy individuals who, during the same period, had gone for a checkup to the medical examination center of the same hospital, were included as controls. All the subjects in this study were Han Chinese females, and the controls were matched according to age (±5 years) and menopausal status.
Ethics statement
The study was approved by the Human Research Committee of Xi’an Jiaotong University. The demographic and personal information of patients and controls was collected using standard epidemiological questionnaires. Clinical information was collected from the patients’ medical and pathological reports. All the participants were informed of the purpose and the experimental procedures of this study, and each subject signed a consent form.
Genotyping assay
Peripheral blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-coated tubes and were preserved at -80°C.31 Genomic DNA was extracted from whole blood samples using the Universal Genomic DNA Extraction Kit (version 3.0; TaKaRa Bio Inc., Kusatsu, Japan) according to the manufacturer’s instructions. DNA concentration was determined using the DU530 UV/VIS spectrophotometer (Beckman Instruments, Fullerton, CA, USA).32,33 Data from the HapMap database were used to create a list of potentially functional Fas and FasL SNPs discovered in Chinese subjects. Only SNPs with a minor allele frequency of >0.01 were considered, in order to ensure a statistical power of at least 50%. In the end, two SNPs were chosen to be included in this study, Fas rs1800682 and FasL. A multiplexed SNP MassEXTEND assay was designed using the MassARRAY Assay Design 3.0 Software (Agena Bioscience, San Diego, CA, USA). All the SNPs were genotyped by the Sequenom MassARRAY RS1000 system (Agena Bioscience), according to the manufacturer’s instructions. The primers used for the two SNPs are listed in Table 1. Data were analyzed using the Typer 3.0 Software (Agena Bioscience).
  | Table 1 Primers used for this study |
Statistical analysis
We used Microsoft Excel for data management and SPSS software (version 21.0, IBM Corporation, Armonk, NY, USA) for statistical analysis. Hardy–Weinberg equilibrium was examined using Fisher’s exact test. Two-tailed Pearson’s chi-square tests were used to evaluate the differences in allelic frequencies for each SNP between patients and controls, with P-values <0.05 considered statistically significant. Five different genetic models were used to evaluate the association between SNPs and breast cancer risk (“A” and “a” are used to symbolize the major and the minor alleles, respectively): the allele model (a vs A); the codominant model (homozygote model: aa vs AA; heterozygote model: Aa vs AA); the recessive model (aa vs AA+Aa); the dominant model (AA vs Aa+aa); and the overdominant model (AA+aa vs Aa). SPSS software was used to estimate ORs and 95% confidence intervals (CIs) for each model. Stratification analysis was used to adjust for possible cofounders. Power and Sample Size (PS) Calculation software (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) was used to calculate the power of the significant difference.34 Haplotype analysis was performed using PHASE v 2.1 software, while ORs and 95% CIs for all the haplotypes were determined using SPSS.35,36
Results
Characteristics of patients and controls
As shown in Table 2, a total of 1,143 subjects (560 cancer cases and 583 healthy controls) were included in this study. Sporadic breast cancer patients were recruited from the Department of Oncology of The Second Affiliated Hospital of Xi’an Jiaotong University. Their average age was 49.09±11.02 years, and the diagnosis was confirmed by the examination of the pathology of surgical specimens. The controls we recruited were matched according to age and menopausal status. As a result, there was no significant difference between cases and controls with regard to general characteristics (P>0.05). However, patients and controls significantly differed in the body mass index (kg/m2) value (P=0.038), which indicates that body mass index may be a confounding variable. Therefore, all the results were adjusted to body mass index.
Associations between Fas/FasL SNPs and the risk of breast cancer
The genotype frequency distribution of Fas rs1800682 and FasL rs763110 is shown in Table 3. The genotype distributions in the control group were all in Hardy–Weinberg equilibrium (P-values of 0.163 and 0.817, respectively). Further analysis showed that significant differences exist among rs1800682 and rs763110 alleles with respect to breast cancer risk. The Fas rs1800682 G allele associated with high risk of breast cancer in the heterozygote, dominant, and overdominant models (AG vs AA: OR =1.37, 95% CI =1.06–1.78, P=0.015; AG+GG vs AA: OR =1.32, 95% CI =1.03–1.68, P=0.027; AA+AG vs GG: OR =1.32, 95% CI =1.04–1.66, P=0.021), with power values of 0.964, 0.856, and 0.910, respectively. In contrast, the rs763110 polymorphism associated with reduced risk of breast cancer (homozygote model: OR =0.63, 95% CI =0.40–0.98, P=0.040; dominant model: OR =0.76, 95% CI =0.60–0.96, P=0.022; allele model: OR =0.79, 95% CI =0.66–0.95, P=0.013). Moreover, power calculations confirm that the sample size was large enough to discover the differences among cases and controls in rs763110 (power =0.834, 0.906, and 0.719, respectively).
Stratification analysis of the association of Fas/FasL SNPs with breast cancer risk by age and menopausal status
The subgroup analyses were conducted in order to reveal the influence of age and menopausal status on breast cancer risk. A stratified analysis by age revealed significant associations between the two SNPs and the risk of breast cancer in females >49 years of age in the allele model (rs1800682: OR =1.29, 95% CI =1.01–1.66, P=0.041; rs763110: OR =0.76, 95% CI =0.59–0.99, P=0.040) (Table 4). Subgroup analyses by menopausal status found no association between rs1800682 and breast cancer risk in any genetic model (all, P>0.05) (Table 5). However, it was discovered that the rs763110 polymorphism was associated with lower breast cancer risk among postmenopausal females (heterozygote model: OR =0.69, 95%CI =0.49–0.97, P=0.030; dominant model: OR =0.70, 95%CI =0.51–0.96, P=0.028).
Associations between Fas/FasL SNPs and the clinicopathological features of breast cancer
We also explored the association between the two SNPs and the clinicopathological features of breast cancer, namely tumor size, lymph node metastasis, and the expression of the estrogen receptor, the progesterone receptor, the human epidermal growth factor receptor 2 (HER2) and Ki67. No associations were found between rs1800682 and breast cancer risk with respect to any of the aforementioned clinicopathological features, as in all the cases P-values were >0.05 (Table 6). In contrast, it was found that the T allele of rs763110 was associated with a decrease in the incidence of lymph node metastasis among breast cancer patients (allele model: OR =0.75, 95% CI =0.57–0.97, P=0.031), and higher incidence of the HER2-positive type of breast cancer among patients (allele model: OR =1.37, 95% CI =1.03–1.18, P=0.028) (Table 6).
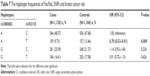
Association between Fas/FasL haplotypes and breast cancer risk
Haplotype analysis was conducted to evaluate the interaction of these two genes. As shown in Table 7, it was found that the haplotype Ars1800682Trs763110 was significantly associated with a lower risk of breast cancer (OR =0.70, 95% CI =0.53–0.91, P=0.009). However, no relation was found between the other two haplotypes (Grs1800682Crs763110 and Grs1800682Trs763110) and breast cancer risk.
Discussion
Apoptosis plays an important role in the genesis and development of tumors. The Fas/FasL genes are key effectors in the regulation of apoptotic cell death, and malfunction of this system has been proved to be important in cancer cell immune evasion and tumorigenesis.37 Decreased expression of Fas promotes malignant transformation and progression, while lower levels of FasL expression have the opposite effect.38–40 Moreover, SNPs of the Fas/FasL genes were reported to be associated with many types of cancer susceptibility. However, the conclusions of these studies were inconsistent and the detailed subgroup analyses were limited. This case–control study including 560 breast cancer cases and 583 controls in Han Chinese females was performed in the hope of providing further evidence regarding the association between Fas/FasL SNPs and cancer risk.
The Fas rs1800682 polymorphism is located in a STAT1 binding element on the Fas promoter, which has been linked to the downregulation of Fas expression.22 The FasL gene rs763110 polymorphism lies within a putative binding motif for the CAAT/enhancer-binding protein β-transcription factor.41 Our results suggest that the Fas rs1800682 SNP may be associated with an increased risk of breast cancer, while the FasL rs763110 polymorphism may decrease breast cancer risk. Furthermore, the associations between these two SNPs and age, menopausal status, and different clinical characteristics of breast cancer were evaluated. Statistically significant associations were found between the two SNPs and the risk of breast cancer in the allele model among patients >49 years of age. Moreover, the rs763110 polymorphism was more likely to decrease breast cancer risk among postmenopausal females. In 2012, a study of the Tunisian population including 438 breast cancer cases and 332 controls found that rs763110 had a marginally significant association with lymph node-negative status (OR =0.53, P=0.06; OR =0.73, P=0.07, respectively).25 However, it is demonstrated that the C allele of the rs763110 SNP may be associated with a lower risk of lymph node metastasis and a higher occurrence of HER2-positive breast cancers. The hormonal status is important to breast cancer patients, guiding treatment and prognosis. It has been demonstrated that upregulation of Fas and FasL in breast cancer cells induces T-cell apoptosis in Fas bearing T cells.42 However, we did not find any associations between Fas/FasL SNPs and hormone-related factors, which is in agreement with the data presented by Crew et al.23 Haplotypes are considered more meaningful than any SNP analysis during the genetic study. Xu et al28 reported that the Grs2234767Ars1800682 haplotype was associated with an increased breast cancer risk as compared with other haplotypes, whereas the Ars2234767Ars1800682 haplotype was associated with a reduced breast cancer risk. However, they did not evaluate the interaction of the Fas and FasL genes. The present study demonstrated that Ars1800682Trs763110 was significantly associated with low risk of breast cancer in comparison with A rs1800682C rs763110. Compared with the previous reports of breast cancer, this study offered more detailed information on these two SNPs and clinical characteristics of breast cancer, which may provide insights into the associations between Fas/FasL polymorphisms and the occurrence and development of breast cancer. Functional studies revealed that rs2234767 and rs1800682 polymorphisms were able to alter the SP1 and STAT1 binding site, leading to the abnormal expression of Fas,43 and C allele of rs763110 could increase basal FasL expression, affecting the FasL-mediated signaling.41 It was found that rs1800682 SNP had a high risk of breast cancer, and the mutation of rs763110 C allele may decrease the breast cancer risk. Thus, it was supposed that G allele of rs1800682 may downregulate Fas transcription via SP1/STAT1 way and the mutation of rs763110 may decrease the FasL expression by regulating FasL-mediated apoptotic signaling. However, the mechanisms still need more functional studies to testify.
Limitations
The study had certain limitations: First, all the subjects were recruited from the same hospital, and therefore, selection bias is inevitable. Second, the sample size was not large enough to support the stratified analyses. Third, the effects of other important risk factors, such as polycyclic aromatic hydrocarbons, exposure of the chest area to high-dose radiation, and alcohol consumption, as well as benign breast lesions or environmental exposures, were not analyzed because of lack of relative data. Fourth, most patients included in this study were invasive ductal carcinoma cases; hence, stratification analyses were not performed for histopathological types of breast cancers. Fifth, only the differences of gene distribution in the subjects were analyzed. Functional studies are needed to reveal the molecular mechanism. Thus, a study with more patients and controls, containing both gene distribution and functional data, would allow us to both confirm and mechanistically interpret the results presented in the current study.
Conclusion
In conclusion, this case–control study indicates that the Fas rs1800683 SNP is associated with a statistically significant degree of increased breast cancer risk, especially in females >49 years of age. Conversely, the FasL rs763110 polymorphism was shown to be associated with decreased risk of breast cancer, especially in females >49 years of age and after menopause. Moreover, stratified analyses demonstrated that the FasL rs763110 SNP was related to reducing the incidence of lymph node metastasis and increased chance for the breast cancer to be HER2-positive. Furthermore, the Ars1800682Trs763110 haplotype may play a protective role against breast cancer.
Acknowledgments
We thank Dr Tian Feng, Hong-Tao Ren, and Yu-Yao Zhu for data selecting and statistical analysis. We also thank Editage for the language editing. This study was supported by National Natural Science Foundation, People’s Republic of China (No 81471670; 81274136); China Postdoctoral Science Foundation (No 2014M560791); the Fundamental Research Funds for the Central Universities, People’s Republic of China (No 2014qngz-04); the International Cooperative Project (No 2013KW-32-01), and Science and Technology Plan of Innovation Project, Shaanxi Province, People’s Republic of China (No 2015KTCL03-06).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. | ||
Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. | ||
Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. | ||
Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. | ||
Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. | ||
Nagata S, Golstein P. The Fas death factor. Science. 1995;267(5203):1449–1456. | ||
Kaufmann T, Strasser A, Jost PJ. Fas death receptor signalling: roles of Bid and XIAP. Cell Death Differ. 2012;19(1):42–50. | ||
Kim JG, Sohn SK, Chae YS, et al. TP53 codon 72 polymorphism associated with prognosis in patients with advanced gastric cancer treated with paclitaxel and cisplatin. Cancer Chemother Pharmacol. 2009;64(2):355–360. | ||
Hagleitner MM, Coenen MJ, Gelderblom H, et al. A first step toward personalized medicine in osteosarcoma: pharmacogenetics as predictive marker of outcome after chemotherapy-based treatment. Clin Cancer Res. 2015;21(15):3436–3441. | ||
Kang S, Dong SM, Seo SS, Kim JW, Park SY. FAS-1377 G/A polymorphism and the risk of lymph node metastasis in cervical cancer. Cancer Genet Cytogenet. 2008;180(1):1–5. | ||
Kordi Tamandani DM, Sobti RC, Shekari M. Association of Fas-670 gene polymorphism with risk of cervical cancer in North Indian population. Clin Exp Obstet Gynecol. 2008;35(3):183–186. | ||
Zucchi F, da Silva ID, Ribalta JC, et al. Fas/CD95 promoter polymorphism gene and its relationship with cervical carcinoma. Eur J Gynaecol Oncol. 2009;30(2):142–144. | ||
Tan SC. Letter regarding “CD95 rs1800682 polymorphism and cervical cancer risk: evidence from a meta-analysis” by Zhang et al. Tumour Biol. 2015;36(11):8275–8276. | ||
Cao Y, Miao XP, Huang MY, et al. Polymorphisms of death pathway genes FAS and FASL and risk of nasopharyngeal carcinoma. Mol Carcinog. 2010;49(11):944–950. | ||
Zhu Q, Wang T, Ren J, Hu K, Liu W, Wu G. FAS-670A/G polymorphism: a biomarker for the metastasis of nasopharyngeal carcinoma in a Chinese population. Clin Chim Acta. 2010;411(3–4):179–183. | ||
Zhang F, Sturgis EM, Sun Y, et al. Apoptotic variants as predictors of risk of oropharyngeal cancer recurrence after definitive radiotherapy. Int J Cancer. 2015;137(10):2454–2461. | ||
Yang M, Sun T, Wang L, et al. Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res. 2008;14(10):3230–3236. | ||
Zhang J, Liu Q, Mao HT. [Analyzing of Fas-670 gene polymorphism in hepatocarcinoma tissue]. Zhonghua gan zang bing za zhi. 2009;17(8):630–631. Chinese. | ||
Zhou RM, Wang N, Chen ZF, Duan YN, Sun DL, Li Y. Polymorphisms in promoter region of FAS and FASL gene and risk of cardia gastric adenocarcinoma. J Gastroenterol Hepatol. 2010;25(3):555–561. | ||
Bye H, Prescott NJ, Matejcic M, et al. Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis. 2011;32(12):1855–1861. | ||
Wang S, Wu S, Meng Q, et al. FAS rs2234767 and rs1800682 polymorphisms jointly contributed to risk of colorectal cancer by affecting SP1/STAT1 complex recruitment to chromatin. Sci Rep. 2016;6:19229. | ||
Crew KD, Gammon MD, Terry MB, et al. Genetic polymorphisms in the apoptosis-associated genes FAS and FASL and breast cancer risk. Carcinogenesis. 2007;28(12):2548–2551. | ||
Zhang B, Sun T, Xue L, et al. Functional polymorphisms in FAS and FASL contribute to increased apoptosis of tumor infiltration lymphocytes and risk of breast cancer. Carcinogenesis. 2007;28(5):1067–1073. | ||
Mahfoudh W, Bouaouina N, Gabbouj S, Chouchane L. FASL-844 T/C polymorphism: a biomarker of good prognosis of breast cancer in the Tunisian population. Hum Immunol. 2012;73(9):932–938. | ||
Wang W, Zheng Z, Yu W, Lin H, Cui B, Cao F. Polymorphisms of the FAS and FASL genes and risk of breast cancer. Oncol Lett. 2012;3(3):625–628. | ||
Hashemi M, Fazaeli A, Ghavami S, et al. Functional polymorphisms of FAS and FASL gene and risk of breast cancer – pilot study of 134 cases. PLoS One. 2013;8(1):e53075. | ||
Xu Y, Deng Q, He B, et al. The diplotype Fas-1377A/-670G as a genetic marker to predict a lower risk of breast cancer in Chinese women. Tumour Biol. 2014;35(9):9147–9161. | ||
Cheng J, Zhou T, Liu C, et al. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263(5154):1759–1762. | ||
Sheen-Chen SM, Chen HS, Eng HL, Chen WJ. Circulating soluble Fas in patients with breast cancer. World J Surg. 2003;27(1):10–13. | ||
Dai ZJ, Liu XH, Kang HF, et al. Genetic variation in metastasis-associated in colon cancer-1 and the risk of breast cancer among the Chinese Han population: a STROBE-Compliant Observational Study. Medicine. 2016;95(6):e2801. | ||
Dai ZJ, Liu XH, Ma YF, et al. Association between single nucleotide polymorphisms in DNA polymerase kappa gene and breast cancer risk in Chinese Han population: a STROBE-Compliant Observational Study. Medicine (Baltimore). 2016;95(2):e2466. | ||
Wang M, Wang X, Fu SW, et al. Single-nucleotide polymorphisms in PSCA and the risk of breast cancer in a Chinese population. Oncotarget. Epub 2016 Mar 30. | ||
Dupont WD, Plummer WD Jr. Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19(6):589–601. | ||
Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. | ||
Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. | ||
Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 1993;75(6):1169–1178. | ||
Griffith TS, Brunner T, Fletcher SM, Green DR, Ferguson TA. Fas ligand-induced apoptosis as a mechanism of immune privilege. Science. 1995;270(5239):1189–1192. | ||
Lee SH, Shin MS, Park WS, et al. Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene. 1999;18(25):3754–3760. | ||
Reichmann E. The biological role of the Fas/FasL system during tumor formation and progression. Semin Cancer Biol. 2002;12(4):309–315. | ||
Wu J, Metz C, Xu X, et al. A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol. 2003;170(1):132–138. | ||
O’Connell J, Bennett MW, O’Sullivan GC, O’Callaghan J, Collins JK, Shanahan F. Expression of Fas (CD95/APO-1) ligand by human breast cancers: significance for tumor immune privilege. Clin Diagn Lab Immunol. 1999;6(4):457–463. | ||
Sibley K, Rollinson S, Allan JM, et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63(15):4327–4330. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.