Back to Archived Journals » Current Biomarker Findings » Volume 6
Differential prognostic values of mRNA expression of CEACAM gene family members in nonsmall cell lung cancer
Authors Nakamura H, Sakai H, Miyazawa T, Somehara T, Nakayama N, Fujii K, Nishimura T
Received 1 August 2016
Accepted for publication 15 October 2016
Published 25 November 2016 Volume 2016:6 Pages 23—30
DOI https://doi.org/10.2147/CBF.S118627
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Hung Khong
Haruhiko Nakamura,1,2 Hiroki Sakai,1 Tomoyuki Miyazawa,1 Toshiaki Somehara,2 Noboru Nakayama,2 Kiyonaga Fujii,2 Toshihide Nishimura2
1Department of Chest Surgery, 2Department of Translational Medicine Informatics, St Marianna University School of Medicine, Kawasaki, Kanagawa, Japan
Abstract: Serum carcinoembryonic antigen (CEA) is widely used as a representative marker of various malignant tumors. CEA-related cell adhesion molecules (CEACAMs), including CEACAM5, are encoded in the human genome by 12 independent genes and can be potential targets for future cancer treatments. In nonsmall cell lung cancer, serum CEA levels have been reported to predict patient survival. However, associations between mRNA expression of CEACAM gene family members in tumor tissues and patient prognosis remain unclear. To clarify this point, we used the Kaplan–Meier plotter global portal site, which collects the results of Affymetrix gene expression microarray analyses from the publicly accessible Gene Expression Omnibus database and combined it with survival data of patients. A total of 1,926 nonsmall cell lung cancer patients were identified from the Gene Expression Omnibus series, Cancer Biomedical Informatics Grid, and The Cancer Genome Atlas databases. We found statistically significant associations between mRNA expression of several CEACAMs and overall survival (OS) in patients with nonsmall cell lung cancer and lung adenocarcinoma (n=720) but not squamous cell carcinoma (n=524). In adenocarcinoma, higher expression levels of CEACAM6 and CEACAM8 were significantly associated with better OS, whereas higher expression levels of CEACAM3, CEACAM4, CEACAM19, and CEACAM21 were associated with worse OS. Conflicting results among multiple probe sets for the same gene were found for CEACAM1, CEACAM5, and CEACAM7. The findings of this study indicated that CEACAMs play important roles in tumor progression and impact OS of patients with adenocarcinoma. As the impact on OS differed based on the gene family members or the probe set used, the individual CEACAMs seem to function through complicated mechanisms. Further studies are necessary to resolve the problems encountered in our present study.
Keywords: mRNA, microarray, survival, nonsmall cell lung cancer, CEACAM, CEA
Introduction
Approximately 50 years ago, carcinoembryonic antigen (CEA) was identified as an oncofetal antigen in colorectal cancer in addition to normal human fetal organs, including the gut, liver, and pancreas.1 Further studies revealed the presence of CEA and numerous CEA cross-reacting antigens in human sera, normal tissues, and various cancers other than colorectal cancer.2–5 A family of CEA-related cell adhesion molecules (CEACAMs), including CEACAM5, is known to be encoded in the human genome by 12 independent genes on chromosome 19q13.2,6,7 These CEACAM proteins belong to the immunoglobulin supergene family, and the molecules contain one or two variable-like domains with or without constant 2-like domains.2 CEA is widely used as a representative serum tumor marker of various malignant tumors and elevated serum CEA levels are reported to be frequently associated with a poor clinical outcome in cancer patients presumably through a variety of mechanisms, including the promotion of invasion, dissemination, metastasis, and immune suppression.2 Serum CEA concentrations are increased in both nonsmall cell lung cancer (NSCLC) and small cell lung cancer.8–10 In NSCLC, elevated serum CEA levels have been reported to be associated with histological types, advanced disease stages, and worse prognoses.8,11–15 More recently, vaccination therapy, antibody therapy, and small interfering RNA therapy targeting CEACAMs have been developed as new therapies for several solid tumors, including lung cancer.16–22 Thus, CEA and related molecules are important not only for diagnoses but also for future therapeutic targets in various malignant tumors.23
In spite of many reports regarding associations of serum CEA levels and prognosis of NSCLC, information about mRNA expression in tumor tissues and its relationship to patient survival is quite limited. Thus, we studied associations between mRNA expression detected by gene expression microarrays and overall survival (OS) in NSCLC patients by accessing an online public database.24,25 To the best of our knowledge, this is the first report focusing on associations between mRNA expression of the CEACAM gene family members and OS in NSCLC patients.
Materials and methods
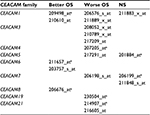
Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository supported by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Based on the GEO database for NSCLC, the Kaplan–Meier plotter global portal site (http://kmplot.com/analysis/index.php?p=service&cancer=lung) provides combined data of mRNA expression of independent genes on gene expression microarrays and survival data of patients with NSCLC.24 Microarray platforms in the provided database were Affymetrix HG-U133A (GPL96), HG-U133 Plus 2.0 (GPL570), and HG-U133A 2.0 (GPL3921). These microarrays have 22,277 probe sets in common.24 A total of 1,926 NSCLC patients were identified from the GEO series, Cancer Biomedical Informatics Grid (https://github.com/NCIP/caarray; caArray project), and The Cancer Genome Atlas (http://cancergenome.nih.gov) database. The data sets used in the present study are shown in Table 1.26–37 Characteristics of the CEACAM gene family members are listed in Table 2.2,6,7 Associations between mRNA expression profiles of nine CEACAM genes (CEACAM1, CEACAM3, CEACAM4, CEACAM5, CEACAM6, CEACAM7, CEACAM8, CEACAM19, and CEACAM21) in tumor tissues and OS of NSCLC patients were studied. Among the CEACAM genes, three (CEACAM16, CEACAM18, and CEACAM20) were not included in the studied microarrays.
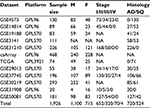  | Table 1 Datasets of nonsmall cell lung cancer included in the analysis Abbreviations: AD, adenocarcinoma; F, females; M, males; NA, not applicable; SQ, squamous cell carcinoma. |
On the Affymetrix gene expression microarrays, six CEACAM genes were detected by multiple different probe sets: five probes for CEACAM1, three for CEACAM3 and CEACAM7, and two for CEACAM5, CEACAM6, and CEACAM21. A single reliable probe set matching each of the examined genes was determined by assessing specificity, splice isoform coverage, and robustness against transcript degradation. These selected probes are marked as Jetset probes38 on the webpage.
The OS rates of two groups of patients subdivided by the median value of mRNA expression were calculated, and Kaplan–Meier survival plots were obtained from the webpage. Simultaneously, the hazard ratio of the higher expression group relative to the lower expression group, 95% confidence intervals, and log-rank p-values were automatically calculated on the same webpage.
Results
Differences in OS between groups with higher and lower expression levels of the investigated CEACAM genes are shown according to histological types: NSCLC, adenocarcinoma (AD), and squamous cell carcinoma (SQ; Table 3). Expression of most CEACAM family members was significantly associated with OS in patients with NSCLC (n=1,926). Similar results were obtained for patients with AD (n=720). In contrast, none of the examined CEACAM gene family members were associated with OS in patients with SQ (n=524), suggesting that the significant differences in OS among NSCLC patients were mainly due to OS differences in AD patients.
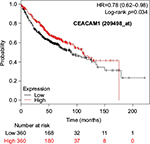
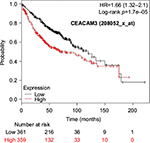
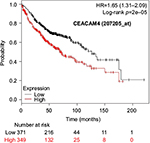
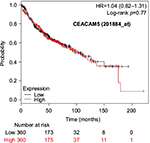
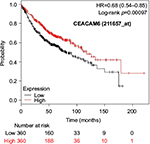
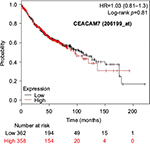
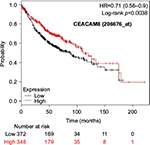
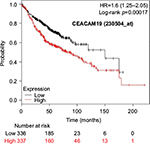
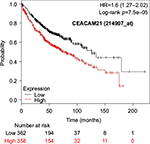
The prognostic value of mRNA expression of CEACAM genes in AD tissues according to the individual probe sets of the microarrays are summarized in Table 4. Higher expression of CEACAM6 and CEACAM8 was associated with better OS, whereas higher expression of CEACAM3, CEACAM4, CEACAM19, and CEACAM21 was associated with worse OS. Conflicting results among multiple probe sets for the same gene were found in CEACAM1, CEACAM5, and CEACAM7.
Kaplan–Meier plots of OS using the Jetset probes for individual genes are shown in Figures 1 9. Because the online database listed no Jetset probe for CEACAM3, 208052_x_at was used as a representative probe for this gene to construct the OS curves.
Discussion
Quantification of serum CEA in lung cancer patients is widely performed to arrive at a diagnosis, evaluate tumor responses to various therapeutic modalities, and predict risks of postsurgical recurrences. However, evidence of prognostic values in lung cancer patients remains unclear. Because of this, there are no official guidelines or recommendations for the use of CEA as a prognostic indicator of lung cancer.
According to a recent review article regarding the prognostic significance of CEA in lung cancer, 18 studies reported statistically significant evidence for the use of CEA as a prognostic marker in NSCLC patients, while seven studies showed negative results.39 Among the 25 studies included in this review article, only one examined the relationship between immunohistochemical CEA expression in tumor tissues and prognoses of patients but found no association.40 A meta-analysis of 16 studies (4,296 NSCLC patients) reported that preoperative high serum CEA levels were associated with poor OS with a combined hazard ratio of 2.28.41 This meta-analysis concluded that preoperative serum CEA levels can predict OS in patients with NSCLC, although high heterogeneity between included studies and publication biases should be taken into consideration.
Little is known about the functions of CEACAMs, particularly impacts on lung cancer tumorigenesis and development. However, the functions and prognostic values of CEACAM1, CEACAM5, and CEACAM6 have been investigated in several tumors. For instance, surface expression of CEACAM1-4L in A549 human lung AD cells is reported to play a critical role in differentiation, contact-inhibited cell growth, and tumor suppressive functions.42 Disturbances in CEACAM1-4L signaling in A549 cells by CEACAM1-4S and other CEACAMs, such as CEACAM5 and CEACAM6, lead to undifferentiated cell growth and malignant transformation. In contrast, multiple clinical studies reported that CEACAM1 overexpression was associated with worse prognosis in melanoma,43 gastric cancer,44 thyroid cancer,45 and NSCLC,46 suggesting that CEACAM1 contributes to tumor progression. Thus, there are discrepancies concerning the functions of CEACAM1 among these studies. In colorectal cancer, immunohistochemically detected CEACAM6 overexpression in tumor tissues independently predicted poor OS and shortened disease-free survival, whereas CEACAM1 and CEACAM5 were not significantly related to these outcomes.47 In epidermal growth factor receptor mutation-negative lung AD patients, a immunohistochemical study of tumor tissues revealed that CEACAM6 expression was associated with worse prognoses, whereas CEACAM3 expression was associated with better prognoses.48 These studies indicated that CEACAM6 overexpression was a worse prognostic factor for selected lung cancer patients.
In the present study, none of the CEACAM gene family members were predictive of OS in patients with SQ. The most apparent result in this study is that CEACAM expression in lung SQ is not useful to predict OS. In contrast, statistically significant differences in OS were confirmed in NSCLC and AD. Since NSCLC is mainly composed of AD and SQ, differences in OS among NSCLC patients are mostly a reflection of OS differences in AD patients.
Associations with worse OS in patients with AD were confirmed by higher mRNA expression of CEACAM3, CEACAM4, CEACAM19, and CEACAM21, while CEACAM6 and CEACAM8 were associated with better OS. Since CEACAM6 overexpression is reportedly associated with worse prognosis of various cancers,47–51 the results of the present study are unique and should be confirmed in further studies. We found no reports examining serum concentration or expression levels of the other CEACAMs and associated impacts on survival of cancer patients.
For some CEACAM gene family members, conflicting results were obtained because of the use of unique probes for each gene. This is not surprising because a given gene may be detected by multiple probe sets on an Affymetrix microarray, which can result in inconsistent or even contradictory findings. The cross-reactivity of probes to other genes and multiple transcripts produced by alternative splicing events are plausible reasons. In order to create simple one-to-one mapping between genes and probe sets, a scoring system using a specific algorithm was proposed for Jetset probes as the most reliable, and these probes are identified on the Kaplan–Meier plotter webpage.38 The most conflicting results were found for the CEACAM1 probe sets. Because CEACAM1 has 13 splice variants and each may have a different function in lung cancer progression,42 different prognostic significance might be due to the different specificity of the probe set reactive with the different variants. Jetset probe 209498_at for CEACAM1 indicated better OS. Similarly, conflicting results were found in CEACAM5 and CEACAM7. No differences in OS based on mRNA expression were found for Jetset probe sets 201884_at for CEACAM5 and 206199_at for CEACAM7.
Since there is quite limited information concerning the relationships between overexpressed CEACAMs in tumor tissues and survival of lung cancer patients, data mining using an accessible public database is both reasonable and useful. A major limitation to this study was the limited clinical information of individual patients, thus it was difficult to perform subgroup analyses. However, the total number of included patients was sufficient to obtain reliable results, if interpreted cautiously.
In conclusion, we found statistically significant associations between mRNA expression of CEACAMs and OS of NSCLC patients. These close associations were confirmed in AD, but not in SQ, suggesting that CEACAMs play important roles in progression of AD. Since the impact of expression of individual CEACAMs on OS differed (better, worse, or neutral) based on the gene family members or used probe sets, each CEACAM seems to function through complicated mechanisms. Hence, further studies are necessary to resolve the problems encountered in the present study.
Disclosure
The authors report no conflicts of interest in this work.
References
Gold P, Freedman SO. Demonstration of tumor-specific antigens in human colonic carcinoma by immunological tolerance and absorption techniques. J Exp Med. 1965;121:439–462. | ||
Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(3–4):643–671. | ||
Chevinsky AH. CEA in tumors of other than colorectal origin. Semin Surg Oncol. 1991;7(3):162–166. | ||
Hasegawa T, Isobe K, Tsuchiya Y, Oikawa S, Nakazato H, Nakashima I, Shimokata K. Nonspecific crossreacting antigen (NCA) is a major member of the carcinoembryonic antigen (CEA)-related gene family expressed in lung cancer. Br J Cancer. 1993;67(1):58–65. | ||
Allard WJ, Neaman IE, Elting JJ, Barnett TR, Yoshimura H, Fritsche HA, Yeung KK. Nonspecific cross-antigen 50/90 is elevated in patients with breast, lung, and colon cancer. Cancer Res. 1994;54(5):1227–1234. | ||
Tchoupa AK, Schuhmacher T, Hauck CR. Signaling by epithelial members of the CEACAM family – mucosal docking sites for pathogenic bacteria. Cell Commun Signal. 2014;12:27. | ||
Pavlopoulou A, Scorilas A. A comprehensive phylogenetic and structural analysis of the carcinoembryonic antigen (CEA) gene family. Genome Biol Evol. 2014;6(6):1314–1326. | ||
Vincent RG, Chu TM, Fergen TB, Ostrander M. Carcinoembryonic antigen in 228 patients with carcinoma of the lung. Cancer. 1975;36(6):2069–2076. | ||
Goslin RH, Skarin AT, Zamcheck N. Carcinoembryonic antigen. A useful monitor of therapy of small cell lung cancer. JAMA. 1981;246(19):2173–2176. | ||
Lokich JJ. Plasma CEA levels in small cell lung cancer. Correlation with stage, distribution of metastases, and survival. Cancer. 1982;50(10):2154–2156. | ||
Hsu WH, Huang CS, Hsu HS, Huang WJ, Lee HC, Huang BS, Huang MH. Preoperative serum carcinoembryonic antigen level is a prognostic factor in women with early non-small-cell lung cancer. Ann Thorac Surg. 2007;83(2):419–424. | ||
Matsuoka K, Sumitomo S, Nakashima N, Nakajima D, Misaki N. Prognostic value of carcinoembryonic antigen and CYFRA21-1 in patients with pathological stage I non-small cell lung cancer. Eur J Cardiothorac Surg. 2007;32(3):435–439. | ||
Matsuguma H, Nakahara R, Igarashi S, et al. Pathologic stage I non-small cell lung cancer with high levels of preoperative serum carcinoembryonic antigen: clinicopathologic characteristics and prognosis. J Thorac Cardiovasc Surg. 2008;135(1):44–49. | ||
Tomita M, Shimizu T, Hara M, Ayabe T, Onitsuka T. Serum carcinoembryonic antigen level in non-small-cell lung cancer patients with preoperative normal serum level. Gen Thorac Cardiovasc Surg. 2009;57(6):303–306. | ||
Nakamura H, Saji H, Marushima H, Kimura H, Koizumi H, Takagi M. Associations between serum carcinoembryonic antigen levels and adenocarcinoma subtypes of the lung. Cancer Treat Commun. 2016;3:31–35. | ||
Berinstein NL. Carcinoembryonic antigen as a target for therapeutic anticancer vaccines: a review. J Clin Oncol. 2002;20(8):2197–2207. | ||
McCann KJ, Mander A, Cazaly A, et al. Targeting carcinoembryonic antigen with DNA vaccination: on-target adverse events link with immunological and clinical outcomes. Clin Cancer Res. 2016;22(19):4827–4836. | ||
Aurisicchio L, Roscilli G, Marra E, Luberto L, Mancini R, La Monica N, Ciliberto G. Superior immunologic and therapeutic efficacy of a xenogeneic genetic cancer vaccine targeting carcinoembryonic human antigen. Hum Gene Ther. 2015;26(6):386–398. | ||
Staff C, Magnusson CG, Hojjat-Farsangi M, et al. Induction of IgM, IgA and IgE antibodies in colorectal cancer patients vaccinated with a recombinant CEA protein. J Clin Immunol. 2012;32(4):855–865. | ||
Hong KP, Shin MH, Yoon S, et al. Therapeutic effect of anti CEACAM6 monoclonal antibody against lung adenocarcinoma by enhancing anoikis sensitivity. Biomaterials. 2015;67:32–41. | ||
Molina-Pinelo S, Gutierrez G, Pastor MD, et al. MicroRNA-dependent regulation of transcription in non-small cell lung cancer. PLoS One. 2014;9(3):e90524. | ||
Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Ann. Surg. 2004;240(4):667–674; discussion 675–676. | ||
Turriziani M, Fantini M, Benvenuto M, et al. Carcinoembryonic antigen (CEA)-based cancer vaccines: recent patents and antitumor effects from experimental models to clinical trials. Recent Pat Anticancer Drug Discov. 2012;7(3):265–296. | ||
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. | ||
You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Des Devel Ther. 2015;9:5087–5097. | ||
Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–7472. | ||
Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28(29):4417–4424. | ||
Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. | ||
Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–37. | ||
Yamauchi M, Yamaguchi R, Nakata A, et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One. 2012;7(9):e43923. | ||
Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. | ||
Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R; Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. | ||
Xie Y, Xiao G, Coombes KR, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17(17):5705–5714. | ||
Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19(1):194–204. | ||
Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5(186):186ra66. | ||
Girard L, Rodriguez-Canales J, Behrens C, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res. 2016;22(19):4880–4889. | ||
Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. Thorac Oncol. 2014;9(1):59–64. | ||
Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. | ||
Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76(2):138–143. | ||
Ford CH, Stokes HJ, Newman CE. Carcinoembryonic antigen and prognosis after radical surgery for lung cancer: immunocytochemical localization and serum levels. Br J Cancer. 1981;44(2):145–153. | ||
Wang XB, Li J, Han Y. Prognostic significance of preoperative serum carcinoembryonic antigen in non-small cell lung cancer: a meta-analysis. Tumour Biol. 2014;35(10):10105–10110. | ||
Singer BB, Scheffrahn I, Kammerer R, Suttorp N, Ergun S, Slevogt H. Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS One. 2010;5(1):e8747. | ||
Thies A, Moll I, Berger J, et al. CEACAM1 expression in cutaneous malignant melanoma predicts the development of metastatic disease. J Clin Oncol. 2002;20(10):2530–2536. | ||
Zhou CJ, Liu B, Zhu KX, et al. The different expression of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) and possible roles in gastric carcinomas. Pathol Res Pract. 2009;205(7):483–489. | ||
Liu W, Wei W, Winer D, et al. CEACAM1 impedes thyroid cancer growth but promotes invasiveness: a putative mechanism for early metastases. Oncogene. 2007;26(19):2747–2758. | ||
Laack E, Nikbakht H, Peters A, et al. Expression of CEACAM1 in adenocarcinoma of the lung: a factor of independent prognostic significance. J Clin Oncol. 2002;20(21):4279–4284. | ||
Jantscheff P, Terracciano L, Lowy A, et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol. 2003;21(19):3638–3646. | ||
Kobayashi M, Miki Y, Ebina M, et al. Carcinoembryonic antigen-related cell adhesion molecules as surrogate markers for EGFR inhibitor sensitivity in human lung adenocarcinoma. Br J Cancer. 2012;107(10):1745–1753. | ||
Kim KS, Kim JT, Lee SJ, et al. Overexpression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in colorectal cancer. Clin Chim Acta. 2013;415:12–19. | ||
Chen J, Li Q, An Y, et al. CEACAM6 induces epithelial-mesenchymal transition and mediates invasion and metastasis in pancreatic cancer. Int J Oncol. 2013;43(3):877–885. | ||
Zang M, Zhang B, Zhang Y, et al. CEACAM6 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition via PI3K/AKT signaling pathway. PLoS One. 2014;9(11):e112908. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.