Back to Archived Journals » Energy and Emission Control Technologies » Volume 3
Consolidated bioprocessing for biofuel production: recent advances
Received 13 December 2014
Accepted for publication 12 January 2015
Published 6 May 2015 Volume 2015:3 Pages 23—44
DOI https://doi.org/10.2147/EECT.S63000
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adolfo Perujo
Veronica Mbaneme-Smith, Mari S Chinn
Department of Biological and Agricultural Engineering, North Carolina State University, Raleigh, NC, USA
Abstract: Consolidated bioprocessing (CBP), the combination of saccharolytic enzyme production and secretion, hydrolysis of polysaccharides, and fermentation of available sugars within a single-unit operation, improves cellulose conversion efficiency and decreases lignocellulosic biomass processing costs for producing biofuels and value-added products. Clostridium thermocellum, an anaerobic, thermophilic bacterium is a significant biocatalyst aspirant for CBP, due to its native cellulolytic and ethanologenic capabilities. This review highlights strain development/modification, metabolic engineering, and process improvements associated with CBP in the context of using C. thermocellum as a model biocatalyst to reduce operating expenditures and inhibitory effects for enhanced biofuels production. In addition, opportunities in using a microbial consortia and biofilms are discussed. As an overview of recent advances in CBP technologies to convert lignocellulosic biomass to biofuels, this review gives researchers a platform for future development of efficient and sustainable CBP approaches.
Keywords: Clostridium thermocellum, cellulosome, anaerobic, cellulolytic, ethanol
Introduction
Expected population growth with corresponding energy usage, limiting resources, and environmental stresses will require development of sustainable technological advances to meet future energy demands that do not compromise available resources.1 Sustainable developments, such as renewable energy systems, equalize energy supply and demand while improving food, economic, energy, and environmental security.2 In 2012, 19% of the global final energy consumption came from renewable forms of energy, with 10.2% (50.3 EJ yr−1) supplied from various forms of biomass.3,4 Lignocellulosic biomass, organic biological matter derived from agricultural, municipal, and forestry operations, is a renewable resource that can be used for the production of reliable forms of energy, such as biofuels.5,6 The attractiveness of using lignocellulosic biomass as raw material in biofuel production processes is attributed to abundance, relatively low expense, and environmentally negligible production and potential for global distribution.7,8 While lignocellulosics possess roughly 40%–50% cellulose, 20%–40% hemicellulose, and 20%–30% lignin by weight, the major challenge is the presence of lignin, effective breakdown of the recalcitrant cellulose structure, and efficient bioconversion to liquid fuels.9,10 The conversion of biomass through biochemical techniques, including pretreatment, hydrolysis, and fermentation, has the potential to produce heat, electricity, fiber, chemicals, and biofuels while improving economic and environmental sustainability.11 To this end, significant research has focused on consolidated bioprocessing (CBP), a conversion approach, though still in its infancy stages of development, that combines all rate-limiting processes, pretreatment, saccharification, and fermentation, within a single reactor to decrease operating cost, increase conversion efficiencies, and reduce by-product inhibition. While a plethora of microbes possess cellulolytic and solventogenic capabilities, Clostridium thermocellum is a strong candidate for CBP, due to its natural ability to rapidly solubilize cellulose by way of its cellulosome, a multi-enzyme complex, and produce ethanol. Additionally, C. thermocellum is an anaerobic thermophile, which eliminates the need for aeration, decreases contamination concerns, and improves temperature-dependent functionality of hydrolytic enzymes.12–14 This review focuses on highlighting process improvements, recombinant microbial catalyst and enzyme development, and metabolic engineering associated with CBP in the context of using C. thermocellum as a model biocatalyst and providing an overview in support of further research in sustainable biofuel production from lignocellulosic biomass. In addition, recent opportunities in using microbial consortia and biofilms are discussed.
Lignocellulosic biomass structure
Lignocellulose comprises of three main polymers, cellulose, hemicelluloses, and lignin, linked in a dense complex matrix with varying compositions depending on type, species, and source of biomass, in addition to pectin, extractives, and structural ash.15–18 Cellulose, the main component of the plant cell wall bestowing structural support, is composed of β-d-glucopyranose moieties linked through β-(1,4) glycosidic bonds.7,8 While disaccharide cellobiose is the repeating unit of the cellulose chain, the degree of polymerization in nature ranges from 2,000 to 20,000 units, and when combined through covalent and hydrogen bonding as well as Van der Waals forces form microfibrils.15,18,19 Hydrogen bonding within a cellulose microfibril determines stability or straightness of the chain, while hydrogen bonds between cellulosic units introduce order, such as crystalline structures, or disorder with amorphous structures.20 In addition, due to the intra- and interchain hydrogen bonding, cellulose is insoluble in most solvents, including water, and considered extremely resistant to enzymatic hydrolysis.7 The cellulose microfibril structures are integrated with hemicellulose, various noncrystalline sugars, and other polymers, such as pectin. Hemicellulose is the second most abundant polymer and contrasts with cellulose by lacking chemical homogeneity, possessing a lower molecular weight and short lateral chains that are relatively easily hydrolyzed.8,15,18,21 While differing in composition, hemicelluloses are highly branched, heterogeneous polymers of pentoses, such as xylose and arabinose; hexoses, such as mannose, glucose, and galactose; and acetylated sugars.8,22,23 Among the main polymers in lignocellulosic materials, hemicellulose is the most thermochemically susceptible and alleged to cover the cellulose microfibrils; therefore, to significantly increase cellulose digestibility, at least 50% of this polymer must be removed.24 Lignin, the adhesive binding various components of lignocellulosic biomass together, is a highly complex three-dimensional amorphous heteropolymer network of phenyl propane units, such as p-coumaryl, coniferyl, and sinapyl alcohol, which provides strength as well as opposition to microbial attack and oxidative stress for plant cell walls.24,25 While lignin is a valuable coproduct for the generation of heat, power, and solid fuel, the noncarbohydrate polymer creates a major limitation in enzymatic and microbial hydrolysis of lignocellulosic biomass.7,26 Chang and Holtzapple27 showed that biomass digestibility was enhanced with increasing lignin removal and in addition to being a physical barrier, the disadvantageous effects of lignin consisted of nonspecific adsorption of hydrolytic enzymes, interference with, and nonproductive binding of cellulolytic enzymes to lignin–carbohydrate complexes, and toxicity of lignin derivatives to microorganisms. Determining the comparative copiousness of cellulose, hemicellulose, and lignin polymers is essential for selecting optimal energy conversion systems to reduce processing time and cost.10,28–30
Advancements in bioprocessing
During the past decade, technological advances have promoted commercial feasibility of biochemical conversion processing for the production of liquid fuels from lignocellulosic biomass.2 This renewable energy production approach includes such operations as biomass size reduction, pretreatment, hydrolysis, and fermentation to produce value-added products.31 Size reduction or comminution mechanically lacerates biomass to facilitate enzymatic accessibility to lignocellulosic materials for further pretreatment and digestibility.32 Pretreatment strategies strive to enhance biomass porosity to increase sugar yields during subsequent hydrolysis and degradation failure of pentoses, as well as to improve the breakdown and recovery of lignin- and hemicellulose-derived sugars, while minimizing by-product inhibition of later unit operations.8,33 Following pretreatment of lignocellulosic biomass, hydrolysis, also known as saccharification, converts complex carbohydrates into simple five- and six-carbon monomers, such as arabinose, fructose, galactose, glucose, mannose, and xylose, by using acids or highly specific external sources of cellulases and hemicellulases.34–36 In contrast with acid hydrolysis, enzymatic hydrolysis has advantages in reduced energy requirements and imperturbable environments without the formation of inhibitory byproducts.37 Furthermore, enzymatic hydrolysis has proven to be advantageous, due to nominal toxicity, decreased utility expenditures, and truncated corrosion compared to acid or alkaline hydrolysis.38–42 Subsequent to saccharification, fermentation procedures can be conducted as batch, fed-batch, continuous, and solid-substrate processes depending on the kinetic properties of choice microbes, source of lignocellulosic hydrolyzate, and process economics to generate biofuels.43,44 Extensive understanding of the genomes, physiologies, and metabolisms of Saccharomyces cerevisiae, Zymomonas mobilis, and Escherichia coli has resulted in their industrial utilization in bioconversion operations.45,46 Natively, S. cerevisiae is incompetent in the fermentation of pentose sugars, such as xylose and arabinose, nor proficient at functioning at optimal exogenous hydrolytic enzymatic temperatures between 50°C and 60°C.5,45 Therefore, metabolic and evolutionary engineering was required to enhance usability of various hydrolyzed monomers and tolerance to environmental stressors.47 Generally, strain engineering of saccharolytic and fermentative microbes are conducted to express cellulases and hemicellulases, secrete proteins, produce biofuels, and tolerate solvents and lignin byproducts, such as furans, weak organic acids, and phenolics, from fermentation procedures.48
Separate hydrolysis and fermentation and simultaneous saccharification and fermentation are two bioconversion technologies generally commissioned for commercial implementation (Figures 1A and 1B).16 Enzymatic hydrolysis can also be performed simultaneously with the cofermentation of glucose and xylose as separate operations in a process known as simultaneous saccharification and cofermentation (Figure 1C).47 Saccharification and fermentation and saccharification and cofermentation have advantages over traditional separate hydrolysis and fermentation, often supporting increased alcohol yields by eradicating by-product inhibition and improved rates of hydrolysis, as well as reduced cost of operation by jettisoning the need for a secondary reactor, lowering enzyme loadings, and decreasing processing time and risk of contamination.48,49 Technological advancements to improve operating procedures and biomass solubilization effectiveness naturally led to the holistic CBP approach consisting of three biotransformations.48 To achieve CBP benefits, cellulase generation, saccharification of biomass, and fermentation of reducing sugars take place in conjunction by a monoculture or coculture of microorganisms, such as bacteria like C. thermocellum and fungi including Neurospora crassa, Fusarium oxysporum, S. cerevisiae, and Paecilomyces sp., for the generation of biofuels (eg, ethanol, butanol) and other value-added products.12,35,50–52 Alleviation of production expenditures and reduced chemical usage can be achieved by CBP, as the microbes used reduce the need for a multitude of external enzymes, which are not substrate specific.16,48 Additionally, this approach decreases the need for numerous reactors and offers simplicity in operating procedures by minimizing the compatibility requirements of enzyme and fermentation systems.53–55 In fact, a comparative cost analysis conducted on ethanol production, when considering capital, raw materials, utilities, and yield loss expenditures, resulted in a projection of $0.04 gal−1 for CBP, while a saccharification and cofermentation was projected at $0.19 gal−1.54 Eclectic selections of microorganisms have been reconnoitered for CBP (Table 1); however, a microbial consortium that combines optimum substrate use and product formation properties, such as rapid hydrolysis rates and enhanced product concentrations and selectivity is preferable in CBP systems.56 In addition, anaerobic metabolism is imperative to CBP, due to superfluous expenditures associated with aerating large culture volumes, as well as product and lignocellulosic energy inefficiencies, as a result of aerobic respiration.13 Thermophilic microorganisms are also necessary to improve hydrolysis rates, which is a major challenge when conducting CBP at optimum ambient conditions for mesophilic microbes of interest.48 Although, the potential and rationale for CBP of biofuels exists, long fermentation periods and meager biofuel yields resulting from by-product formation (eg, organic acids), sensitivity of microorganisms to alcoholic solvents, and limited growth in hydrolysis supernatant resulted in significant research focused on improving the robustness of microorganisms utilized in CBP by employing recombinant strategies on cellulolytic and solventogenic organisms.57,58 While biochemical conversion systems demonstrate prodigious potentials for producing biofuels with minimal environmental impacts, the present challenge is efficiently converting lignocellulosic material to available sugars and fermenting crude sugars to biofuels with a robust microorganism and relatively high operating costs.59,60
  | Table 1 Cellulolytic and hemicellulolytic, ethanologenic (including genetically modified) microorganisms utilized for consolidated bioprocessing |
Native cellulolytic and ethanologenic microbes
C. thermocellum, an extensively researched strict anaerobic, acetogenic, cellulolytic, and thermophilic bacterium, possesses the ability to degrade cellulose and ferment mixed products from cellulose solubilization, while producing ethanol, acetate, hydrogen (H2), carbon dioxide (CO2), lactate, and formate under various growth conditions.61–67 Cellulose degradation rates approaching 2.5 g L−1 h−1, thermally stable biomass degrading enzymes, and proclivity to synthesize value-added products makes C. thermocellum of great interest to CBP development.61,68,69 In addition, an optimum growth temperature of 60°C precludes that of most contaminating microorganisms, which allows for C. thermocellum cultures to be maintained and viable when using nonsterile substrates for pure-culture fermentation processes.70 High fermentation temperatures and anaerobic conditions also reduce operational expenditures for ethanol recovery and reactor cooling, which are typically associated with fermentations using mesophilic, aerobic bacterium.68 Decreases in solubility of product gases (H2 and CO2) at higher fermentation temperatures also allow enhanced removal efficiency of product gases to be achieved, based on Henry’s law.71 The ability to hydrolyze most recalcitrant forms of cellulose as effectively as the commercially significant fungus, Trichoderma reesei, in combination with the robustness of thermostable enzymes, limited contamination and continuous distillation of fermentation products at elevated temperatures has resulted in the persistent interest in C. thermocellum as a biocatalyst in CBP for ethanol generation (Table 2).72
Fermentation by C. thermocellum
C. thermocellum hydrolyzes cellulosic substrates into shorter chain sugars, primarily cellobiose for conversion to ethanol, acetate, and lactate as primary end products through various metabolic pathways (Figure 2).73 Instead of a phosphotransferase system, C. thermocellum mediates the transportation of cellobiose using an ATP-binding cassette.74–76 Phosphorylation of cellobiose and glucose to glucose 1-phosphate takes place prior to conversion through glycolysis as the predominate pathway, with less common flux through the pentose phosphate pathway or the Entner–Doudoroff pathway.64,73,76 Most of the pyruvate is converted to acetyl-CoA, reduced Fd (FdH2), and CO2 with a significant amount abridged to lactate by lactate dehydrogenase.77 Subsequently, reduction of acetyl-CoA to acetaldehyde and then to ethanol catalyzed by NAD+-linked dehydrogenase transpires or acetate is formed yielding stoichiometric amounts of ATP.77,78 Utilizing the three-branched pathway to metabolize hexoses without genetic manipulation or the addition of external factors, C. thermocellum has been shown to produce 0.6 mol to 1.0 mol of ethanol per mol of hexose with an ethanol tolerance of 5 g L−1, due to a high degree of membrane fluidity based on lipid content.77,79,80 Aside from ethanol, acetate, and lactate, other compounds including pyruvate, malate, uracil, soluble glucan, and free amino acids, which normally are products disassociated with C. thermocellum, have been observed in the liquid fraction of lignocellulosic fermentations.81
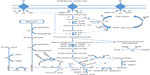  | Figure 2 Metabolic functionality of Clostridium thermocellum cellulosome for product formation. |
Varying substrate complexity and cultivation systems render a multitude of product ratios for a particular strain (Table 2), in addition to versatile functionality and inherent by-product tolerance. For instance, S14 advantageously possesses the capability to cultivate in an inclusive temperature and pH growth range and produce considerable quantities of cellulosomal enzymes in the medium.82 Furthermore, cellulosomes of S14 were shown to have an increased activity on microcrystalline cellulose and lignocellulosics, such as rice straw, with higher resistance to cellobiose inhibition, compared to the cellulosomes of ATCC 27405.82
In addition to substrate utilization, supplemental nutrients, such as salts, vitamins, and reducing agents, are of great importance to product formation when utilizing C. thermocellum and other anaerobes.83,84 Islam et al85 demonstrated enhanced cellulose fermentation and end-product synthesis by C. thermocellum DSM 1237 with varied nutrient compositions of α-cellulose, yeast extract, urea, CaCl2 · 2H2O, MgCl2 · 6H2O, FeSO · 6H2O, and vitamins under carbon-excess conditions. The researchers also illustrated two major effects, a general growth enhancement effect and a carbon flux shifting effect, which rendered volumetric ethanol and H2 yields that increased by 2.3-fold and 2.04-fold, respectively, when compared to the basic nutritional blend.
Shifting metabolic product synthesis in C. thermocellum can also be induced with the supplement of acetone, ethanol, formate, or sodium azide or by increasing the hydrostatic bioreactor pressure by incorporating exogenous gases.14,16,86,87 More specifically, the addition of acetate when initiating fermentative procedures increases ethanol and decreases formate production, while the addition of ethanol increases hydrogen and acetate concentrations. Formate proves advantageous not only at increasing hydrogen and ethanol but also at decreasing acetate. Alternatively, the initial incorporation of hydrogen was determined to decrease carbon dioxide and increase formate production, while the addition of headspace carbon monoxide considerably enhanced the production of ethanol and inhibited the generation of H2, CO2, and acetate.16 Shifts in end-product formation can also be accomplished through strain modifications, such as with the evolved M1570 (Δhpt, Δldh, Δpta), which produced a 40:1 molar ratio of ethanol to organic acid while making 5.61 g L−1 ethanol.69
Cellulosome structure and capability
C. thermocellum expresses a collection of cellulolytic enzymes, which accumulate into large, complex, multi-protein structures on the cell surface, known as cellulosomes.70,88 Cellulosomes with molecular weights ranging from 2.0 to 6.5 × 106 and encompassing 14–26 polypeptide subunits, depending on strain and growth conditions, were first recognized while conducting research on adhesion factors linking bacterium to cellulose.89–99 Specifically, an electron micrograph demonstrated that the cytoplasmic membrane of C. thermocellum was bordered by a thin peptidoglycan layer, a regular S-layer, and an amorphous outer layer formed by the protuberate cellulosome.68,100 By way of the cellulosome, C. thermocellum indeed attaches to cellulose, while over 20 enzymes, including endoglucanases, cellobiohydrolases, and xylanases, within the cellulosome efficiently degrade cellulose to glucose and cellulodextrins, followed by subsequent transportation for cellular metabolism, in earlier stages of cellulolysis.71,101 More specifically, during optimal growth conditions, C. thermocellum forms a monolayer biofilm without an extracellular polymeric matrix, parallel to the carbon fibers of the lignocellulosic substrate, caricaturing the topography to facilitate extracellular hydrolysis.102,103 During the latter stages of a cell culture, it has been reported that the cellulosome detaches from the bacterium cell; however, the possibility exists that the cellulosome remains attached to high-molecular weight cellulose compounds.96 The effectiveness of cellulosomes to catalyze the hydrolysis of cellulose depends significantly on the maintenance of structural integrity, as partially disassociated cellulosome complexes result in major activity loss, especially as it relates to crystalline forms of cellulose.104 Nonetheless, economic production of cellulolytic enzymes and reducing enzyme to biomass ratio required for commercialization of biofuels derived from lignocellulosic biomass could be achieved when utilizing cellulolytic CBP microbes for enzyme production, secretion, and recycling.104,105 While advancements are still required in enzyme recycling, solutions can be found exploiting the tethering capability of the C. thermocellum cellulosome.12,105
Definitively, the primary and largest structural subunit of the multi-functional C. thermocellum cellulosome, known as the cellulosome-integrating protein (CipA), is a glycoprotein with a mass of 210–250 kDa and functions as a nonhydrolytic scaffoldin.94,98 The modular protein consists of a carbohydrate-binding module (CBM-3a), also referred to as the cellulose-binding domain, with a comprehensive binding specificity for crystalline cellulose, a hydrophilic module of unknown functionality, termed X, a modified dockerin domain and nine internal repeated sequences or hydrophobic cohesion domains positioned relative to the amino terminus, which bind catalytic subunits.106,107 Specifically, amalgamation of the catalytic subunits into the cellulosome transpires by way of calcium-dependent interactions between the scaffoldin cohesions and Type I dockerin domains of the catalytic subunits.108,109 Furthermore, the cellulosome binds to cellulose via the CBM-3a, as well as through the CBMs of the catalytic subunits. In fact, Kataeva et al110 demonstrated synergism between the catalytic subunit, CelD, and the CBM of the C. thermocellum scaffoldin. Type II dockerin domains attached to the C-terminus of the bacterium scaffoldin do not bind to the scaffoldin cohesion domains, instead, these dockerin domains bind to specific Type II cohesion domains on the cell surface, known as anchoring proteins.111–114 These proteins (SdbA, Orf2p, and OlpB) contain an S-layer homology module for correlating with the cell surface and consequently anchoring the cellulosome to the cell.115 While 23 cellobiohydrolase (exoglucanases) genes for cellulosomal components that contain dockerin domains have been recognized, endoglucanases have also been ascertained, as well as lichenase, chitinase, mannanase, and five xylanases, with two containing xylan esterase modules that remove feruloyl residues.104,116–120 Although C. thermocellum is incapable of fermenting xylan, xylose, and other five-carbon sugars, the latter enzymes are believed to breakdown glycans encasing cellulose microfibrils to improve accessibility of the cellulose to be degraded by cellulosomes.115
The efficient degradation of lignocellulosic biomass is due to the juxtaposition of directing the enzymatic complex to the substrate, known as the targeting effect, and the spatial propinquity of the different types of cellulases to one another, known as the proximity effect.121 While the cellulosome assembles in a temperately selective manner, this multi-enzyme system is one of the most efficient natural biocatalyst for degrading lignocellulosic biomass, therefore research groups have attempted to capture its function physically, incorporate it into other microorganisms, and mimic its behavior in synthesized enzyme complexes.109,122–124
Recombinant cellulolytic approach
Microbial strain engineering to facilitate high cellulolytic activity, the ability to use numerous carbon sources, and resistance to toxic compounds released during the pretreatment of lignocellulosic biomass are important for the development of cost-effective fermentation of lignocellulosics for the directed generation of biofuels and value-added chemicals.125 To this end, recombinant cellulolytic approaches originating from natural anaerobic mesophilic and thermophilic bacteria and fungi possessing cellulosomes (Table 3) have been investigated.117,126,127 In fact, natural cellulosomes detectible on the cell surface increases the localized concentration of enzymes allowing for enhanced consumption of cellobiose and cellodextrins by a microorganism, which minimizes inhibition related to product formation.88 Through the use of electron microscopy, Fourier-transform infrared spectroscopy, and X-ray diffraction analysis, synergistic enzymatic activity of the cellulosomes are considered advantageous over aerobic bacteria and fungi, which exhibit fluctuating enzymatic functionality and specificity associated with external enzyme secretion.16,128 C. thermocellum is the most cellulolytic thermophile and distinctively possesses exceptionality with regard to size, complexity, and genomic arrangement of its cellulosome, vindicating one of the highest rates of cellulose degradation and utilization.14,88,117,129,130 As a result, the recombination of cellulosomal components of C. thermocellum into minicellulosomes has been engineered onto the surface of Aspergillus niger, Bacillus subtilis, Clostridium acetobutylicum, E. coli, Lactococcus lactis, Thermoanaerobacterium saccharolyticum, and S. cerevisiae by way of anchoring proteins or cohesion–dockerin interactions.5,131–137
  | Table 3 Studied bacteria and fungi with cellulosomal structures |
Engineering of bacteria
With regard to bacteria, fragments of the scaffoldin protein CipA from C. thermocellum containing a single cohesin module, two cohesin modules, one cohesin and a cellulose-binding module, or only a cellulose-binding module were functionally displayed on the cell surface of L. lactis.134,135 To alleviate challenges associated with cell toxicity from protein overexpression, a nisA inducible promoter was used as well as the C-terminal-anchoring streptococcal M6 motif.135 While significant variation in efficiencies were exhibited among the designed minicellulosomes, accredited to structural characteristics in protein conformation, scaffold size, and presence of noncohesion modules, the surface display of functional scaffold proteins proved imperative for developing recombinant microorganisms capable of using multiple metabolic pathways to directly convert carbonaceous biomass into biofuels and other value-added chemicals.135 Similarly, B. subtilis was engineered to display multiple thermophilic cellulase enzymes (Cel8A) from C. thermocellum on the cell surface by way of proteins containing a staphylococcus aureus cell wall sorting signal covalently anchored to peptidoglycan by coexpression with the Bacillus anthracis sortase A (SrtA) transpeptidase.136 Additionally, a Cel8A–dockerin fusion protein was stably anchored onto B. subtilis utilizing noncovalent cohesion–dockerin interactions to increase lignocellulosic degradation potential during CBP.136
The capability of T. saccharolyticum to unhesitatingly solubilize hemicellulose with the heterologous expression of a functional cellulosome makes this Gram-positive anaerobic thermophile of interest in CBP. In fact, expression and localization of a full-length CipA from C. thermocellum was observed by developing an inducible system based on the native xynA T. saccharolyticum promoter, regulated by xylan and xylose.131 As part of the study, an exorbitant quantity of xylose and SigmaCell 101, a microcrystalline cellulose, was hydrolyzed when the ΔcipA mutant C. thermocellum was cocultured with the CipA-expressing T. saccharolyticum strain.131
In addition to native ethanologenic species, other solventogenic microbes, such as C. acetobutylicum, that proficiently convert sugars to ethanol and butanol are of interest in CBP; however, these species are known to inefficiently grow on and metabolize crystalline cellulose, which has prompted the engineering of secretion and assemblage of cellulosomes by allele-coupled exchange technology.133 Specifically, BioBrick2-standardized fragments were used to assemble a range of synthetic genes encoding C. thermocellum cellulosomal scaffoldin proteins (CipA variants) and glycoside hydrolases (GHs, Cel8A, Cel9B, Cel48S, and Cel9K) as well as synthetic cellulosomal operons that direct the synthesis of Cel8A, Cel9B, and a truncated form of CipA. Successful heterologous protein expression, secretion, and self-assembly of cellulosomal subunits by recombinant C. acetobutylicum was demonstrated by utilizing supported allele-coupled exchange technology as a platform for synthesizing novel cellulosomes for CBP.
Engineering of fungi
A. niger was utilized in CBP, as a direct result of its inherent ability to secrete feruloyl esterases that hydrolyze diferulate cross-links in lignocellulose and enhance overall hydrolysis performance.137 In an effort to facilitate a stronger synergism between catalytic domains, a minicellulosome was incorporated into A. niger to prevent large-scale diffusion of enzymes and lower protein requirements for hydrolysis. In detail, a chimeric protein composed of the feruloyl esterase A from A. niger was associated with the Cel48S dockerin from C. thermocellum and produced in A. niger. Analyzing the chimeric enzyme for its binding capacity indicated that translational fusion to glucoamylase improved the secretion efficiency of the protein and allowed production of the first functional fungal enzyme joined to a bacterial dockerin.137
For S. cerevisiae, most cell surface display methods developed have been based on agglutinin and flocculin model systems, which incorporate cell wall proteins (α-agglutinin, Aga1, Cwp1, Cwp2, Tip1p, Srp1, Flo1p, Sed1p, Tir1p, and YCR89W) containing a glycosylphosphatidylinositol (GPI) signal motif covalently cross-linked to β-1,6-glucan.138 More specifically, S. cerevisiae was genetically modified to assemble a cell surface designer cellulosome by heterologously expressing a chimeric scaffoldin protein (Scaf3p) regulated by a phosphoglycerate kinase 1 promoter, β-xylanase 2 secretion signal, cell wall protein 2 (Cwp2), and termination sequences, for GPI-mediated anchoring to the cell wall.138 While fluorescent microscopy confirmed that Scaf3p targeted the yeast cell surface, Far Western blot analysis demonstrated functionality of the C. thermocellum dockerin domain binding the Scaf3 protein. Phenotypic evidence for cohesin–dockerin interaction was also established with the detection of a two-fold increase in tethered endoglucanase enzyme activity in S. cerevisiae cells compared to the wild type. This work highlighted the feasibility of designing cellulolytic strains of S. cerevisiae through emulation of the cellulosome concept.
Tsai et al139 functionally displayed a cell surface minicellulosome exhibiting three divergent cohesin domains (CelE, CelA, and CelG) from C. thermocellum, Clostridium cellulolyticum, and Ruminococcus flavefaciens on S. cerevisiae. The recombinant cellulosome maintained synergistically significant glucose liberation and produced ethanol when utilizing a β-glucosidase (BglA) from C. thermocellum tagged with dockerin from R. flavefaciens.139 As a result, S. cerevisiae was engineered to display a succession of uni-, bi-, and trifunctional minicellulosomes consisting of a miniscaffoldin containing a CBM and three cohesion modules tethered to the cell surface through the yeast α-agglutinin adhesion receptor.140 While the cell surface assembly of the minicellulosome was dependent on high-affinity interactions between cohesion–dockerin binding domains, the triplicating enzymatic functionality of the surface-displayed minicellulosome enhanced the capabilities of S. cerevisiae at hydrolyzing lignocellulosic material for ethanol production during CBP by improving enzyme–enzyme and enzyme proximity synergies, as well as conceptually explicating cellulosome synthesis and anatomical modality. Subsequently, a cellulolytic four-strain S. cerevisiae consortium capable of either displaying a CipA scaffoldin derived from C. thermocellum or secreting one of three types of cellulases, CelA from C. thermocellum, CBHII from T. reesei, or BGLI from Aspergillus aculeatus, was developed for CBP.141 A 20% increase in ethanol production was observed by regulating the combination and concentration of the four S. cerevisiae strains, with a CipA:CelA:CBHII:BGLI ratio of 2:3:3:0.53 producing 1.80 g L−1 ethanol after 94 hours. Advantageously, this system allows for the optimization of ethanol production as well as limitless implementation of enzymes into the minicellulosome construct, such as hemicellulases and pectinases adhered through fusion domains.
Artificial cellulosomes
In an effort to understand the relationship between the cellulosome structure and enzyme activity, reduce enzyme loadings and related costs, and improve compatibility of multi-protein complexes with biorefinery applications for biofuel production, truncated designer cellulosomes were constructed with cohesion modules and proven highly active compared to free cellulases during lignocellulosic degradation.142–144 One such example is the robust 18-subunit, self-assembled synthetic multi-enzymatic complex, known as the rosettazyme, which consists of a CipA cohesion module and four dockerin-containing cellulases (Cel9B, Cel9K, Cel9R, and Cel48S) from C. thermocellum.122,144 More specifically, rosettazymes are thermostable, group II chaperonins (scaffold-base) from the hyperthermo-acidophilic archaeon Sulfolobus shibatae, which in the presence of ATP/Mg2+ assemble into double-ringed structures that truss dockerin-containing endo- and exo-glucanases and enhance the cellulose degradation activity of bound enzymes.144 The versatility of the rosettazymes cohesion–dockerin interaction on the cellulosomal scaffoldin provides a limitless number of arrays for the utilization of fabricating defined enzymatic nanostructures for the production of biofuels.143 A chimeric cohesion-fused β-glucosidase (BglA-CohII) that binds directly to the cellulosome of C. thermocellum through an unoccupied dockerin module of the CBM was designed to direct enzymatic activity to substrate location.145 BglA was proven to increase microcrystalline cellulose and pretreated switchgrass degradation, maintain cellobiose activity, and integrate with the native cellulosome, permitting the C. thermocellum cellulosome to subsist as a homo-oligomer. These findings demonstrate the significance of enzyme targeting to enhance lignocellulosic biomass degradation.
In addition to cohesion–dockerin interaction, conjugation techniques dependent on scaffold material and enzyme type, such as metal affinity between polyhistidine tag and core-shell quantum dots, protein fusion, and zinc-finger protein (ZFP)-guided assembly, have been investigated to improve stability, storage properties, and enzymatic synergies of synthetic multi-protein complexes.122,144 In contrast with rosettazymes, cellulose–protein fusions are single polypeptide chains with cellulose domains internal to the scaffolding protein designed to increase cellulase thermostability and activity.146 Purposefully, CelA from C. thermocellum was inserted into a hyperstable α-helical consensus ankyrin protein domain scaffoldin, which gave rise to an arrangement of multiple cellulose domains at a predetermined spacing within a single polypeptide to optimize reactivity of a repetitive cellulose lattice.146 Nanoparticles can be model supports for cellulose immobilization to enhance enzymatic activity.147 In a study to evaluate the effects of nanoparticles on catalytic performance of artificial cellulosomes, it was found that size was moderately significant and enzyme proximity was extremely important to enhancing enzymatic activity.147 Proteins can also be site-specifically localized onto a double-strand DNA scaffold using DNA-binding proteins, such as ZFPs composed of three subunits recognizing specific 3-bp sequences to create artificial bifunctional cellulosomes for improving the hydrolysis of cellulose.148 By taking advantage of ZFP modality, site-specific docking of CelA and CBM of C. thermocellum onto a single DNA template was achieved.148 As a result of advancements in protein engineering and conjugation technology, artificial cellulosomes have shown great promise by mimicking the enzymatic synergism observed in native cellulosome systems; however, these complexes are still less active than naturally occurring cellulolytic microorganisms.136 While synergistic interactions between CBMs and catalytic domains impact the efficient degradation of lignocellulosic biomass by native, recombinant, and artificial cellulosomal possessing microbes, investigation of molecular mechanisms by way of metabolic fluxes are also required to improve growth potential, uptake and utilization of monosaccharides, alcoholic yields, and resiliency to inhibitory byproducts.149
Genomics, transcriptomics, proteomics, and metabolic fluxes
To understand the complex reactions required to degrade and ferment lignocellulosic biomass to biofuels, significant research has focused on the genomic, transcriptomic, proteomic, and metabolic profiles of C. thermocellum.45 To expand on the knowledge base for clostridial species, such as C. thermocellum, relevant to current biofuel production efforts, the genome was sequenced and shown to include a 3.8-Mb DNA, single-chromosome arrangement consisting of a 39.8% guanine/cytosine content and 3,173 protein-encoding genes.150,151
Microarray transcriptomic analysis on C. thermocellum demonstrated deviations in gene expression levels resulting from modifications in carbon source and structure, nutrient availability, and microbial density, which validates the microbe’s ability to sense and respond to external factors by signaling peptides or transcriptional regulators to transport and metabolize nutrients.152 When altering the fermentative substrate from cellobiose to crystalline cellulose, expression profiles of genes corresponding to energy generation, translation, glycolysis and amino acid, nucleotide, and coenzyme metabolism increased.153 Moreover, expression profiles for cellulosomal genes, inorganic ion transport and metabolism, signal transduction, amino acid transport, phosphate transport, and resistance–nodulation–division transport increased when utilizing pretreated yellow poplar, diluted acid-pretreated Populus, and switchgrass as lignocellulosic carbon sources.154,155 These discoveries highlight the importance of carbon source on the metabolic functionality and performance of C. thermocellum in CBP.
Supporting proteomic studies expressing core metabolism proteins demonstrated growth-phase dependency for positioning of C. thermocellum to efficiently utilize cellulosic substrates and corresponding nutrients, which facilitates specific and consistent protein expression.156 More specifically, metabolic proteins associated with pyruvate synthesis exhibited a decrease in expression, while proteins associated with glycogen metabolism, pyruvate catabolism, and end-product synthesis pathways increased when the microbe transitioned from exponential to stationary phase. Relative expression profiles demonstrated specific proteins utilized in carbohydrate consumption and end-product synthesis, which reinforced previous findings that hydrogen synthesis occurs by way of bifurcating hydrogenases, while ethanol synthesis is principally catalyzed by a bifunctional aldehyde/alcohol dehydrogenase. The differences in expression profiles of core metabolic proteins in response to growth phase may dictate the carbon and electron flux toward energy storage compounds and fermentation products. Cellulosome proteins also illustrate a change in expression as it relates to carbon availability. For instance, hemicellulases, such as XynA, XynC, XynZ, and XghA, are upregulated, in addition to endoglucanases CelA, CelB, CelE, CelG, and GH5, when C. thermocellum is grown on cellobiose; however, when grown on cellulose, GH9 are superiorly expressed with the corresponding surface-anchoring protein OlpB and endoglucanases CelS and CelK.157 The results support the existing theory that expression of scaffoldin-related proteins is coordinately regulated by a catabolite repression mechanism, xylanase expression is prone to a growth rate-independent regulation, and transcriptional control of cellulases, such as endoglucanases, is conditional on catabolite repression. Similar results were demonstrated with pretreated switchgrass with the exception of an increased expression of GH9 and CelK as well as the decrease in xylanases due to the pretreatment-induced reduction in hemicellulose and xylan composition.158
A major challenge in using C. thermocellum for CBP biofuel production is the need to modify the organism for increased production efficiency; however, the process of properly engineering an organism is typically onerous.159 To this end, a genome-scale model of C. thermocellum metabolism, iSR432, was developed by incorporating genomic sequence data, network topology, and experimental measurements of enzyme activities and metabolite fluxes for the generation of a computational apparatus for evaluating the metabolic network of C. thermocellum and facilitating enhanced biofuel production by way of strain engineering.159 In fact, utilization of this model emphasized the correlation among reduction and oxidation states, as well as ethanol secretion, which permitted the prediction of gene deletions and environmental conditions that would potentially increase ethanol production.159
Recombinant solventogenic approach
Engineering to prolong existing metabolic pathways for the production of novel products, accelerating a rate-determining step, engineering enzymatic activities that synthesize unique biocatalytic nanostructures, and shifting metabolic fluxes toward the generation of value-added products, such as biofuels, is of extreme interest for aiding in the commercial viability of C. thermocellum, as a CBP biocatalysts.16 In an effort to improve C. thermocellum ethanol yields by redirecting the carbon flux, a genetic system for making targeted gene knockouts was developed utilizing a toxic uracil analog, 5-fluoroorotic acid to select for deletion of the pyrF gene involved in organic acid production, namely pta, which encodes the enzyme phosphotransacetylase.160 While the C. thermocellum Δpta strain failed to produce acetate, the deletion marginally affected ethanol production.161 Enzymes associated with organic acid formation were also deleted by creating a counter-selection system based on endogenous hpt and T. saccharolyticum tdk genes.69 By combining these selection markers, the use of replicating plasmids to insert and remove markers from the host chromosome was realized.125 Following the deletion of the l-lactate dehydrogenase (ldh) and pta genes from C. thermocellum M1570 with the counter-selection system and 2,000 hours of adaption, the ethanol yield of the mutant increased by 4.2-fold compared to the wild-type strain; however, ethanol yields failed to increase with decreasing acetate and lactate formation.69 Conversely, when using the engineered C. thermocellum M1570 strain, CBP of downregulated caffeic acid 3-O-methyltransferase transgenic switchgrass was realized with a 20% increase in lignocellulosic conversion, followed by the primary production of ethanol, indicating the importance of substrate utilization when developing strains for enhanced biofuel yields.162 In addition, Deng et al61 examined the impact of targeted modification of enzymes associated with the malate shunt pathway in wild-type C. thermocellum DSM1313, including expression of pyruvate kinase gene from T. saccharolyticum, mutation of the phosphoenolpyruvate carboxykinase and deletion of malic enzyme gene to increase ethanol production. The researchers concluded that C. thermocellum with exogenous pyruvate kinase exhibited a 3.25-fold increase in ethanol yield when compared to the wild-type strain. Similarly, when the gene for malic enzyme and part of malate dehydrogenase were deleted, the anaerobic bacterium demonstrated ethanol yields more than three fold higher than that of the wild-type strain.
As a result of solvent toxicity being attributed to chaotropic effects on biological membranes, such as increased fluidity, degraded proteins and RNA, decreased energy generation and nutrient transportation, and damaged DNA and lipids, the development of strains with superior tolerance is imperative for the sustainable production of biofuels with increased yields.134 Using an innovative engineering approach to increase ethanol tolerance, which typically ranges from 10 g L−1 to 20 g L−1 in wild-type strains, as a means of lowering production costs, Shao et al163 isolated ethanol-tolerant strains of C. thermocellum that were capable of growing in medium containing up to 50 g L−1 ethanol. Genomic analysis of the isolated ethanol-tolerant strains revealed six common mutations, which originated in genes associated with ethanol, arginine, and pyrimidine biosynthesis pathways. Cellodextrin synthesis was also shown to be active as well as metabolically balanced.164 Furthermore, Brown et al165 demonstrated that a shift of cofactor specificity from NADH to NADPH for the bifunctional acetaldehyde–CoA/alcohol dehydrogenase enzyme proffers ethanol tolerance to C. thermocellum, resulting from an interference with electron flow. These studies provide knowledge on the mechanisms of ethanol tolerance to further metabolically engineer C. thermocellum aimed at higher ethanol yields to complement increased tolerance; however, the extensive realization necessitates targeted development and optimization of specific characteristics that will assist in the synergistic functionality.45
Microbial consortia
Thus far, recent advances regarding engineered microorganisms to produce a plethora of enzymes and biofuels from a variety of lignocellulosic substrates are associated with low titers and consequently not commercially viable organisms.56 Conversely, CBP conducted utilizing a microbial consortium, which is similar to natural conversion systems, is of interest due to synergies that result in extremely efficient substrate utilization and increased product yields.56 For instance, C. thermocellum demonstrates a high growth rate on crystalline cellulose; nonetheless, limited ethanol yields, failure to effectively metabolize xylan, and exhibiting insufficient growth on xylose or other pentoses have resulted in hemicellulolytic thermophiles being used in conjunction with C. thermocellum to hydrolyze hemicellulose and ferment all sugars present in biomass.125,166
Cocultures with C. thermocellum have been reported with Methanobacterium thermoautotrophicum for methane and acetate as well as Clostridium thermosaccharolyticum for ethanol, butyrate, and acetate, which kinetically illustrated cellulose and cellobiose fermentation with enhanced ethanol yields from corn stover.167 Once Clostridium thermohydrosulfuricum was isolated and discovered to ferment cellobiose, hexoses, and pentoses to ethanol, this too was coupled with C. thermocellum as part of a CBP study.168 Unlike C. thermosaccharolyticum, C. thermohydrosulfuricum fails to produce butyrate and functions at thermophilic temperatures. More specifically, the microbial consortia, C. thermocellum and C. thermohydrosulfuricum, increased ethanol yields by two fold when compared to monocultures, due to the metabolic capability of C. thermocellum’s cellulases to hydrolyze α-cellulose and hemicellulose, enhanced utilization of mono- and disaccharides by C. thermohydrosulfuricum, improved cellulose consumption, increased ethanol production, and decreased acetate production.167 This coculture was also shown to actively ferment MN300 cellulose, Avicel, Solka Floc, SO2-treated wood, and steam-exploded wood.
More recently, Argyros et al69 cocultured C. thermocellum and engineered T. saccharolyticum for 146 hours, which yielded 38.1 g L−1 ethanol from 92.2 g L−1 Avicel, with concentrations of acetic and lactic acids falling below detectable limits. Due to saccharification being crucial in producing lignocellulosic-derived bioethanol during CBP, extreme pH and ethanol concentrations inhibit production efficiency; therefore, several saccharides derived from lignocellulosics were investigated utilizing C. thermocellum and Clostridium thermolacticum.169 The coculture actively fermented glucose, xylose, cellulose, and microcrystallized cellulose; in addition, the alkali environments proved conducive for ethanol production. While fermentation inhibition was observed when conditions exhibited high ethanol concentrations and extreme pH, initially low levels of ethanol resulted in an unforeseen stimulatory influence on ethanol production.169 Conversely, a novel coculture of C. thermocellum and Thermoanaerobacterium aotearoense with pretreated sugarcane bagasse (SCB) under mild alkali conditions for bio-hydrogen production was established, which demonstrated an economically viable and synergetic advantage in bio-hydrogen production over monocultures with untreated SCB.170 This successful microbial consortium is of interest as ethanol is another main product.
In addition to hemicellulolytic thermophiles, mesophiles, such as C. acetobutylicum and Clostridium beijerinckii, have enormous potential in coculturing CBP applications in conjunction with C. thermocellum for n-butanol and ethanol production from lignocellulosics. In fact, alcoholic solvents with more than two carbons, such as butanol, are ideal candidates for alternative fuels, due to compatibility with existing infrastructure, low hygroscopicity, flexible blending ratios and comparable octane value, energy density, and Reid vapor pressure similar to that of gasoline and diesel.48,54,125,171–173 Currently, butanol is utilized for solvent extraction of fats, dye, nitro enamel, plastificator, butyl acetate, phenol formaldehyde resin, and oil-additive manufacturing.174 With these benefits, a sequential coculture approach with C. acetobutylicum and C. thermocellum grown on solka floc or a combination of solka floc and aspen wood xylan was implemented. The results indicated an efficient utilization of all hydrolysis products derived, which produced a 1.7- to 2.6-fold increase in total fermentation products.175 The majority of the fermentation products were acids; however, induction of solventogenesis by butyric acid was suggested as a solution. In regards to C. beijerinckii, a novel strategy for sequential coculturing with C. thermocellum was conducted to increase the production of alcoholic solvents from alkali-extracted corncobs.176 Under combinatory optimal culture conditions for sugar and solvent production, this CBP consortium degraded 88.9 g L−1 of carbonaceous material and produced 19.9 g L−1 of total products, with 10.9 g L−1 ethanol in 200 hours without the addition of butyrate.176 Nonetheless, the application of coculture CBP is vaguely understood with potential challenges of sole acid production and low product titers arising when symbiotically optimizing processing conditions, such as temperature, pH, and nutrient loads, through quorum sensing and biofilm formation for all strains as it relates to control, stability, and productivity.176–178
Biofilms
Comprehensive implementation of microbial biofilms, structured communities immobilized in a matrix of extracellular polymers, has been expansively applied to bioremediation; however, the potential beneficial application of biofilms in CBP for the production of biofuels has not yet been extensively investigated.179–182 Biofilms have the potential to improve lignocellulosic biomass conversion efficiency, due to the concentration of cell-associated hydrolytic enzymes at the biofilm–substrate interface leading to increased reaction rates, a layered microbial structure allowing sequential conversion of complex substrates, and cofermentation of hexose and pentose with corresponding secretion and fungal–bacterial symbioses.183 More impactful is the confined microenvironment within a biofilm selectively remunerating microbes with superior phenotypes deliberated from intercellular gene or signaling convergence, a process which is deficient in suspended cultures, by altering diffusion rates.183,184 In addition, the immobilized property of biofilms, particularly when membrane attached, simplifies the separation of biofuels from microbial producer and liquid media as well as promotes retention of biomass for continued processing. With these benefits and importance of bacterial adherence in microbial lignocellulose conversion, the organization, dynamic formation, and carbon flow associated with biofilms of C. thermocellum was examined using noninvasive, in situ fluorescence imaging.185 Investigation of the biofilm demonstrated the ability to extensively convert lignocellulosic substrates with a characteristic monolayered cell structure without an extracellular polymeric matrix, typically seen in biofilms. Moreover, cell division at the interface and terminal endospores appeared throughout all stages of biofilm growth. While utilizing continuous-flow reactors with an excessively high rate of dilution (2 h−1), biofilm activity under low (44 g L−1) and high (202 g L−1) initial cellulose loadings resulted in fermentative catabolism being comparable, with 4% of metabolized sugar being utilized for cell production.185 The study also observed 75.4% and 66.7% of the low and high cellulose loadings, respectively, being converted to primary carbon metabolites, including ethanol, acetic acid, lactic acid, and carbon dioxide.185 Differences were also observed in ethanol/acetic acid ratios (g/g), suggesting that substrate availability for cell attachment rather than biofilm colonization rates govern the efficiency of cellulose conversion.
The importance of cellulosic surface exposure to microbial hydrolysis has received little attention regardless of implied influence on conversion kinetics; however, spatial heterogeneity of fiber distribution in pure cellulosic sheets made direct measurements of biofilm colonization and surface penetration unattainable.102 This was circumvented by utilizing online measurements of carbon dioxide (CO2) production in continuous-flow reactors, in conjunction with confocal imaging.102 Results illustrated that the specific biofilm development rate of C. thermocellum has a significant effect on overall reactor kinetics during the period of microbial limitation. Due to biomass recalcitrance and the need to simplify enzymatic conversion, a single multi-species biofilm silicone membrane-adhered reactor was designed for CBP that featured both aerobic and anaerobic conditions.56 Concept feasibility was successfully validated by producing ethanol with a 67% yield from un-detoxified whole-slurry dilute acid-pretreated wheat straw by the combined action of T. reesei, S. cerevisiae, and Scheffersomyces stipitis. The results achieved accentuates the potential of the process as a versatile inexpensive sugar platform for holistically producing biofuels and value-added chemicals from lignocellulosic biomass by specifically compiled consortia of industrially proven robust microorganisms in a CBP system.56 Concisely, immobilized biofilms address solvent productivity, inhibitor tolerance, scalability, electron flux, and excessive fermentation periods.
Industries with interest in CBP
While academic advancements have proven beneficial in the promotion of CBP, so have the industrial advancements of Mascoma and Qteros as they relate to a holistic approach for producing biofuels and value-added products from a microbial consortium with C. thermocellum for the economic sustainability of transportation and rural- and agricultural-based sectors. Mascoma Corporation, an innovative renewable-fuels company, was first to report the targeted metabolic engineering of cellulose-fermenting thermophile, C. thermocellum, to reduce the production of unwanted organic acid byproducts and rapidly degrade and metabolize cellulose with high conversion efficiency and tolerance to commercially relevant levels of ethanol.186 To produce high rates, titers, and yields of biofuels and biochemicals from the conversion of starch, sugars, and cellulosics, Mascoma utilizes CBP, the novel breakthrough technology, and high-performing industrial biocatalysts.187 Mascoma has engineered yeast, TransFerm, and TransFerm Yield+ for improved hydrolysis and enhanced xylose fermentation capabilities to increase yields from CBP processes.188 In 2011, Mascoma was awarded $80 million from the Department of Energy to assist in the design, construction, and operation of a commercial-scale hardwood cellulosic ethanol facility in Kinross, MI, USA.189
Qteros, a startup based in Marlborough, MA, USA, is also pursuing CBP with patented Clostridium phytofermentans, an anaerobic microbe with the ability to both convert recalcitrant polysaccharides into available sugars and ferment hydrolyzate into fuel-grade ethanol as the primary fermentation product.190,191 This Q Microbe® genome has been fully sequenced, which revealed characteristics directly relevant to the efficient and cost-effective production of ethanol, such as over 105 different genes responsible for producing lignocellulosic-degrading enzymes regulated by the type of growth substrate utilized. With $100,000 awarded from the Department of Energy, the Q Microbe® has shown consistent saccharification efficiency across a broad range of feedstocks, including wheat straw, sugarcane bagasse, energy crops, such as switchgrass, and agricultural residues, such as corn stover, cob, and fiber.192,193
Conclusion
Biofuel production from lignocellulosic biomass is extremely attractive because of its substantial and renewable availability, relative low cost, and minimal environmental impact. Conventional sugar and starch sources exhibit high theoretical biofuel yields; however, these resources are inadequate for global utilization and impacts on food security. A potential solution lies in agricultural waste and dedicated energy crops that lack human nutritional value and require less land, water, and energy. Furthermore, biofuel production can significantly reduce greenhouse gas emissions. While biochemical conversion systems demonstrate great promise for producing biofuels, challenges of efficiently converting lignocellulosic biomass to available monomers and subsequent fermentation to biofuels with a robust microbe at low operating costs still exist. To resolve limitations in industrial implementation, CBP systems have been explored, with configurations involving C. thermocellum being most widely investigated. CBP has the potential to improve hydrolysis and fermentation efficiencies, eradicate contribution of exogenous saccharolytic enzymes, and eliminate inhibition effects, while requiring low energy with minimal environmental degradation to biologically convert lignocellulosic biomass to biofuels and other value-added chemicals. Hindrance of the overall ability and robustness of biocatalysts observed when incorporating genes from cellulolytic microbes for hydrolytic enzyme functionality has prompted the use of microbial consortia to achieve the advantages of a CBP operation, providing synergies that improve substrate use and increase biofuel yields. Ultimately, the economic production of sufficient quantities of biofuels will require advancements in feedstock development, conversion processes, system integration, marketing, and public policy as it relates to CBP.
Disclosure
The authors report no conflicts of interest in this work.
References
Waldron K. Bioalcohol Production: Biochemical Conversion of Lignocellulosic Biomass. Abington Hall, Granta Park, Great Abington, Cambridge, UK: Woodhead Publishing Limited; 2010. | |
Yilman S, Selim H. A review on the methods for biomass to energy conversion systems design. Renew Sustain Energ Rev. 2013;25:420–430. | |
World Energy Statistics (WRS). International Energy Agency. Paris, France; 2010. | |
Zervos A. Renewables 2014 Global Status Report. Renewable Energy Policy Network for the 21st Century; 2014. | |
Kricka W, Fitzpatrick J, Bond U. Metabolic engineering of yeasts by heterologous enzyme production for generation of cellulose and hemicellulose from biomass: a perspective. Front Microbiol. 2014;5(174):1–11. | |
Serrano-Ruiz J, West R, Dumesic J. Catalytic conversion of renewable biomass resources to fuels and chemicals. Annu Rev Chem Biomol Eng. 2010;1:79–100. | |
Galbe M, Zacchi G. Pretreatment: the key to efficient utilization of lignocellulosic materials. Biomass Bioenergy. 2012;30:1–9. | |
Brodeur G, Yau E, Badal K, Collier J, Ramachandran K, Ramakrishnan S. Chemical and physicochemical pretreatment of lignocellulosic biomass: a review. Enzyme Res. 2011;2011:1–17. | |
Kim T. Pretreatment of lignocellulosic biomass. In: Yang S, El Enshasy H, Thongchul N, Martin Lo Y, editors. Bioprocessing Technologies in Integrated Biorefinery for Production of Biofuels, Biochemicals and Biopolymers from Biomass. New York: Wiley; 2013:488. | |
McKendry P. Energy production from biomass (part 1): overview of biomass. Bioresour Technol. 2002;83:37–46. | |
Cheng J. Biomass to Renewable Energy Processes. Boca Raton, FL, USA: CRC Press/Taylor and Francis Group; 2009. | |
Schuster B, Chinn M. Consolidated bioprocessing of lignocellulosic feedstocks for ethanol fuel production. Bioenerg Res. 2013;6(2):416–435. | |
Olson D, McBride J, Shaw A, Lynd L. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. | |
Lu Y, Zhang Y, Lynd L. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci U S A. 2006;103(44):16165–16169. | |
Agbor V, Cicek N, Sparling R, Berlin A, Levin D. Biomass pretreatment: fundamentals toward applications. Biotechnol Adv. 2011;29:675–685. | |
Carere C, Sparling R, Cicek N, Levin D. Third generation biofuels via direct cellulose fermentation. Int J Mol Sci. 2008;9:1342–1360. | |
Chandra R, Bura R, Mabee W, Berlin A, Pan X, Saddler J. Substrate pretreatment: the key to effective enzymatic hydrolysis of lignocellulosic. Adv Biochem Eng Biotechnol. 2007;108:67–93. | |
Fengel D, Wegener G. Wood Chemistry, Ultrastructure, Reactions. Berlin, New York: Walter de Gruyter; 1984. | |
Desvaux M. Clostridium cellulolyticum: Model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev. 2005;29:741–764. | |
Laureano-Perez L, Teymouri F, Alizadej H, Dale B. Understanding factors that limit enzymatic hydrolysis of biomass: characterization of pretreated corn stover. Appl Biochem Biotechnol. 2005;121(124):1081–1099. | |
Saha B. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30:279–291. | |
Hanson B, Johnson B, Henson R, Riveland N. Seeding rate, seeding depth, and cultivar influence on spring canola performance in the northern Great Plains. Agron J. 2008;100(5):1339–1346. | |
Polari L, Ojansivu P, Mäkelä S, Eckerman C, Holmbom B, Salminen S. Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr Polym. 2002;48(1):29–39. | |
Hendricks A, Zeeman G. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol. 2009;100:10–18. | |
Ralph J, Peng J, Lu F, Hatfield R, Helm R. Are lignins optically active? J Agric Food Chem. 1999;47:2991–2996. | |
Avgerinos G, Wang D. Selective delignification for fermentation of enhancement. Biotechnol Bioeng. 1983;25:67–83. | |
Chang V, Holtzapple M. Fundamental factors affecting biomass enzymatic reactivity. Appl Biochem Biotechnol. 2000;84(86):5–37. | |
Menon V, Rao M. Trends in bioconversion of lignocellulose: biofuels, platform chemicals and biorefinery concept. Prog Energy Combust Sci. 2012;38:522–550. | |
Sheehan J, Camobreco V, Duffield J, Grabowski M, Shapouri H. Life Cycle Inventory of Biodiesel and Petroleum Diesel for use in an Urban Bus. Golden, CO: National Renewable Energy Laboratory; 1998. | |
Helsel Z. In: Fluck R, editor. Energy in Farm Production. New York, NY: Elsevier; 1992:177–201. | |
Houghton J, Weatherwax S, Ferrell J. Breaking the Biological Barriers to Cellulosic Ethanol: A Joint Research Agenda. US Department of Energy, December 7–9, 2005, Rockville, MD; 2006. | |
Palmowski L, Muller J. Influence of the size reduction of organic waste on their anaerobic digestion. In: II International Symposium on Anaerobic Digestion of Solid Waste, Barcelona; June 15–17, 1999; 137–144. | |
Wyman C, Dale B, Elander R, Holtzapple M, Ladisch M, Lee Y. Comparative sugar recovery data from laboratory scale application of leading pretreatment technologies to corn stover. Bioresour Technol. 2005;96:2026–2032. | |
Moniruzzaman Md, Zahangir Alam Md, Sujan S, Hossain M, Jamal M. Enzymatic saccharification of bagasse: effects of different pre-treatment methods. Int J Renewable Energy Res. 2013;3(2):230–234. | |
Sarkar N, Ghosh S, Bannerjee S, Aikat K. Bioethanol production from agricultural wastes: An overview. Renew Energy. 2012;37:19–27. | |
Beguin P, Aubert J. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. | |
Ferreira S, Durate A, Ribeiro M, Queiroz J, Domingues F. Response surface optimization of enzymatic hydrolysis of Cistus ladanifer and Cytisus striatus for bioethanol production. Biochem Eng J. 2009;45:192–200. | |
Taherzadeh M, Karimi K. Enzyme-based hydrolysis process for ethanol from lignocellulosic material: a review. Bioresources. 2007;2(4):707–738. | |
Kitchaiya P, Intanakul P, Yamamoto T. Enhancement of enzymatic hydrolysis of lignocellulosic wastes by microwave pretreatment under atmospheric pressure. J Wood Chem Technol. 2003;23(2):217–225. | |
Park Y, Kang S, Lee J, Hong S, Kim S. Xylanase production in solid-state fermentation by Aspergillus niger mutant using statistical experimental designs. Appl Microbiol Biotechnol. 2002;58:761–766. | |
Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1–11. | |
Duff S, Murray W. Bioconversion of forest products industry waste cellulosics to fuel ethanol: a review. Bioresour Technol. 1996; 55:1–33. | |
Carroll A, Sommerville C. Cellulosic Biofuels. Annu Rev Plant Biol. 2009;60:165–182. | |
Chandel A, Es C, Rudravaram R, Narasu M, Rao L, Ravindra P. Economics and environmental impact of bioethanol production technologies: an appraisal. Biotechnol Mol Biol Rev. 2007;2(1):14–32. | |
Akinosho H, Yee K, Close D, Ragauskas A. The emergence of Clostridium thermocellum as a high utility candidate for consolidate bioprocessing applications. Front Chem. 2014;2(66):1–17. | |
Tracy B, Jones S, Fast A, Indurthi D, Papoutsakis E. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol. 2012;23:364–381. | |
Koppram R, Nielsen F, Albers E, et al. Simultaneous saccharification and co-fermentation for bioethanol production using corncobs at lab, PDU and demo scales. Biotechnol Biofuels. 2013;6:2–12. | |
Parisutham V, Kim T, Lee S. Feasibilities of consolidated bioprocessing microbes: from pretreatment to biofuel production. Bioresour Technol. 2014;161:431–440. | |
Balat M, Balat H, Oz C. Progress in bioethanol processing. Prog Energy Combust Sci. 2008;34:551–573. | |
Bjerre A, Olesen A, Fernqvist T. Pretreatment of wheat straw using combined wet oxidation and alkaline hydrolysis resulting in convertible cellulose and hemicellulose. Biotechnol Bioeng. 1996;49:568–577. | |
Kim S, Baek S, Lee K, Hahn J. Cellulosic ethanol production using yeast consortium displaying a minicellulosome and β-glucosidase. Microb Cell Fact. 2013;12:1–14. | |
Cardona C, Quintero J, Paz I. Production of bioethanol from sugarcane bagasse: status and perspectives. Bioresour Technol. 2013;101(13):4754–4766. | |
Hamelinck C, van Hooijdonk G, Faaij A. Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle-, and long term. Biomass Bioenergy. 2005;28:384–410. | |
Lynd L, van Zyl W, McBride J, Laser M. Consolidated bioprocessing of cellulosic biomass: An update. Curr Opin Biotechnol. 2005;16(5):577–583. | |
Silverstein R. A comparison of chemical pretreatment methods for converting cotton stalks to ethanol, Master’s Thesis (Sharma, R, Advisor), Department of Biological and Agricultural Engineering, NCSU; 2004. | |
Brethauer S, Studer M. Consolidated bioprocessing of lignocellulose by microbial consortium. Energy Environ Sci. 2014;7:1446–1453. | |
Zaldivar J, Nielsen J, Olsson L. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol. 2001;56:17–34. | |
Szczodrak J, Fiedurek J. Technology for conversion of lignocellulosic biomass to ethanol. Biomass Bioenergy. 1996;10:367–375. | |
Foust T, Ibsen K, Dayton D, Hess J, Kenney K. The biorefinery. In: Himmel D, editor. Biomass Recalcitrance: Destructing the Plant Cell Wall for Bioenergy. UK: Wiley-Blackwell; 2008:7–37. | |
Mielenz J. Ethanol production from biomass: technology and commercialization status. Curr Opin Microbiol. 2001;4:324–329. | |
Deng Y, Olson D, Zhou J, Herring C, Shaw A, Lynd L. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng. 2013;15:151–158. | |
Lynd L, Grethlein H, Wolkin R. Fermentation of cellulose substrates in batch and continuous culture by Clostridium thermocellum. Appl Environ Microbiol. 1989;55:3131–3139. | |
Lynd L, Grethlein H. Hydrolysis of dilute acid pretreated hardwood and purified microcrystalline cellulose by cell-free broth from Clostridium thermocellum. Biotechnol Bioeng. 1987;29:92–100. | |
Lamed R, Zeikus G. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanerobium brockii. J Bacteriol. 1980;144:569–578. | |
Ng T, Weimer P, Zeikus J. Cellulolytic and physiological properties of Clostridium thermocellum. Arch Microbiol. 1977;114:1–7. | |
Weimer P, Zeikus G. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence and presence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol. 1977;33:289–297. | |
Patni N, Alexander J. Utilization of glucose by Clostridium thermocellum: presence of glucokinase and other glycolytic enzymes in cell extracts. J Bacteriol. 1971;105:220–225. | |
Yu T, Xu X, Peng Y, Luo Y, Yang K. Cell wall proteome of Clostridium thermocellum and detection of glycoproteins. Microbiol Res. 2012;167:364–371. | |
Argyros D, Tripathi S, Barrett T, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–8294. | |
Demain A, Newcomb M, Wu J. Cellulase, clostridia and ethanol. Microbiol Mol Biol Rev. 2005;69:124–154. | |
Levin D, Islam R, Cicek N, Sparling R. Hydrogen production by Clostridium thermocellum 27405 from cellulosic biomass substrates. Int J Hydrogen Energy. 2006;31:1496–1503. | |
Hazlewood G, Gilbert H. Xylan and cellulose utilization by Clostridia. In: Woods D, editor. The Clostridia and Biotechnology. Stoneham, MA: Butterworth-Heinemann; 1993:313–327. | |
Zhou J, Olson DG, Argyros DA, et al. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol. 2013;79(9):3000–3008. | |
Nataf Y, Yaron S, Stahl F, et al. Cellodextrin and laminaribiose ABC transporters in Clostridium thermocellum. J Bacteriol. 2009;191(1):203–209. | |
Nochur S, Jacobson G, Roberts M, Demain A. Mode of sugar phosphorylation in Clostridium thermocellum. Appl Biochem Biotechnol. 1992;33:33–41. | |
Ng T, Zeikus J. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982;150:1391–1399. | |
Woods. The Clostridia and Biotechnology. Stoneham, MA: Butterworth-Heinemann; 1993. | |
Andreesen J, Bahl H, Gottschalk G. Introduction to the physiology and biochemistry of the genus Clostridium. In: Minton N, Clark D, editors. Biotechnology Handbook. 3rd ed. New York, NY: Plenum Press; 1989:27–62. | |
Timmons M, Knutson B, Nokes S, Strobel H, Lynn B. Analysis of composition and structure of Clostridium thermocellum membranes from wild-type and ethanol-adapted strains. Appl Microbiol Biotechnol. 2009;82:929–939. | |
Herrero A, Gomez R. Development of ethanol tolerance in Clostridium thermocellum: effect of growth temperature. Appl Environ Microbiol. 1980;40:571–577. | |
Ellis L, Holwerda E, Hogsett D, et al. Closing the carbon balance for fermentation by Clostridium thermocellum (ATCC 27405). Bioresour Technol. 2012;103:293–299. | |
Tachaapaikoon C, Kosugi A, Pason P, et al. Isolation and characterization of a new cellulosome-producing Clostridium thermocellum strain. Biodegradation. 2012;23:57–68. | |
Chinn M, Nokes S. Screening of thermophilic anaerobic bacteria for solid state cultivation on lignocellulosic substrates. Biotechnol Prog. 2006;22:53–59. | |
Chinn M, Nokes S, Strobel H. Influence of process conditions on end product formation from Clostridium thermocellum 27405 in solid substrate cultivation on paper pulp sludge. Bioresour Technol. 2007;98:2184–2193. | |
Islam R, Ozmihci S, Cicek N, Sparling R, Levin D. Enhanced cellulose fermentation and end-product synthesis by Clostridium thermocellum with varied nutrient compositions under carbon-excess conditions. Biomass Bioenergy. 2013;48:213–223. | |
Rani K, Swamy M, Seenayya G. Increased ethanol production by metabolic modulation of cellulose fermentation in Clostridium thermocellum. Biotechnol Lett. 1997;19:819–823. | |
Bothun G, Knitson B, Berberich J, Strobel H, Nokes S. Metabolic selectivity and growth of Clostridium thermocellum in continuous culture under elevated hydrostatic pressure. Appl Microbiol Biotechnol. 2004;65:149–157. | |
Lynd L, Weimer P, van Zyl W, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. | |
Doi R, Tamaru Y. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem Rec. 2001;1(1):24–32. | |
Shoham Y, Lamed R, Bayer E. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7(1):275–281. | |
Kohring S, Wiegel J, Mayer F. Subunit composition and glycosidic activities of the cellulose complex from Clostridium thermocellum JW20. Appl Environ Microbiol. 1990;56(12):3798–3804. | |
Lamed R, Bayer E, Millet J. The cellulosome concept: exocellular/extracellular enzyme reactor centers for efficient binding and cellulolysis. In: Aubert J, Beguin P, editors. Biochemistry and Genetics of Cellulose Degradation. FEMS Symposium No 43. London: Academic Press; 1988:101–106. | |
Lamed R, Bayer E. The cellulosome of Clostridium thermocellum. Adv Appl Microbiol. 1988;33:1–46. | |
Wu J, Orme-Johnson W, Demain A. Two components of an extracellular protein aggregate of Clostridium thermocellum together degrade crystalline cellulose. Biochemistry. 1988;27(5):1703–1709. | |
Mayer F, Coughlan M, Mori Y, Ljungdahl L. Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl Environ Microbiol. 1987;53:2785–2792. | |
Bayer E, Lamed R. Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J Bacteriol. 1986;167(3):828–836. | |
Wiegel J, Dykstra M. Clostridium thermocellum: adhesion and sporulation while adhered to cellulose and hemicellulose. Appl Microbiol Biotechnol. 1984;20:59–65. | |
Lamed R, Setter E, Kenig R, Bayer E. The cellulosome – a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various catalytic activities. Biotechnol Bioeng Symp. 1983;13:163–181. | |
Lamed R, Setter E, Bayer E. Characterization of a cellulose-binding, cellulose-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156:818–827. | |
Lemaire M, Miras I, Gounon P, Beguin P. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology. 1998;144:211–217. | |
Wertzj B. Lignocellulosic Biorefineries. Lausanne: EPFL Press; 2013. | |
Dumitrache A, Wolfaardt G, Allen D, Liss S, Lynd L. Tracking the cellulolytic activity of Clostridium thermocellum biofilms. Biotechnol Biofuels. 2013;6:175–190. | |
Zhang Y-H, Lynd L. Regulation of cellulose synthesis in batch and continuous cultures of Clostridium thermocellum. J Bacteriol. 2005;187(1):99–106. | |
Cavka A, Alriksson B, Rose S, van Zyl W, Jonsson L. Production of cellulosic ethanol and enzyme from waste fiber sludge using SSF, recycling of hydrolytic enzymes and yeast a, and recombinant cellulose-producing Aspergillus niger. J Ind Microbiol Biotechnol. 2014;41:1191–1200. | |
Blanch H. Bioprocessing for biofuels. Curr Opin Biotechnol. 2012;23:390–395. | |
Salamitou S, Raynaud O, Lemaire M, Coughlan M, Beguin P, Aubert P. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-integrating protein CipA. J Bacteriol. 1994;176(10):2882–2887. | |
Tokatlidis K, Salamitou S, Beguin P, Dhurjati P, Aubert P. Interaction of the duplicated segments carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 1991;291(2):185–188. | |
Bayer E, Shoham Y, Lamed R. Cellulose-decomposing bacteria and their enzyme systems. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. Vol 2. 3rd ed. New York: Springer; 2000:578–617. | |
Shimon LJ, Bayer EA, Morag E, et al. A cohesion domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly. Structure. 1997;5(3):381–390. | |
Kataeva I, Guglielmi G, Beguin P. Interaction between Clostridium thermocellum endoglucanase CelD and polypeptides derived from the cellulosome-integrating protein CipA. Biochem J. 1997; 326(pt 2):617–624. | |
Leibovitz E, Ohayon H, Gounon P, Béguin P. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J Bacteriol. 1997;179(8):2519–2523. | |
Leibovitz E, Beguin P. A new type of cohesion domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178(11):3077–3084. | |
Lemaire M, Ohayon H, Gounon P, Fujino T, Béguin P. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177(9):2451–2459. | |
Salamitou S, Lemaire M, Fujino T, et al. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J Bacteriol. 1994;176(10):2828–2834. | |
Leschine S. Degradation of polymers: cellulose, xylan, pectin and starch. In: Dürre P, editor. Handbook on Clostridia. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2005:101–131. | |
Zverlov V, Fuchs K, Schwarz W. Chi18A, the endochitinase in the cellulosome of the thermophilic, cellulolytic bacterium Clostridium thermocellum. Appl Environ Microbiol. 2002;68(6):3176–3179. | |
Schwarz W. The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol. 2001;56:634–649. | |
Blum D, Kataeva I, Li X, Ljungdahl L. Feruloyl esterase activity of the Clostridium thermocellum cellulosome can be attributed to previously unknown domains of XynY and XynZ. J Bacteriol. 2000;182(5):1346–1351. | |
Halstead J, Vercoe P, Gilbert H, Davidson K, Hazlewood G. A family 26 mannanase produced by Clostridium thermocellum as a component of the cellulosome contains a domain which is conserved in mannanases from anaerobic fungi. Microbiology. 1999;145(11):3101–3108. | |
Hayashi H, Takehara M, Hattori T, et al. Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum. Appl Microbiol Biotechnol. 1999;51(3):348–357. | |
Fierobe HP, Bayer EA, Tardif C, et al. Degradation of cellulose substrates by cellulosome chimeras: Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. | |
Goncalves G, Mori Y, Kamiya N. Biomolecular assembly strategies to develop potential artificial cellulosomes. Sustainable Chem Processes. 2014;2:1–9. | |
Borne R, Bayer E, Pages S, Perret S, Fierobe H. Unraveling enzyme discrimination during cellulosome assembly independent of cohesion-dockerin affinity. FEBS J. 2013;280:5764–5779. | |
Sakka K, Kishino Y, Sugihara Y, et al. Unusual binding properties of the dockerin module of Clostridium thermocellum endoglucanase CelJ. FEMS Microbiol Letter. 2009;300:249–255. | |
Hasunuma T, Okazaki F, Okai N, Hara K, Ishii J, Kondo A. A review of enzymes and microbes for lignocellulosic biorefinery and the possibility of their application to consolidated bioprocessing technology. Bioresour Technol. 2013;135:513–522. | |
Doi R, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–551. | |
Doi R, Kosugi A, Murashima K, Tamaru Y, Han S. Cellulosomes from mesophilic bacteria. J Bacteriol. 2003;185(20):5907–5914. | |
Ding SY, Xu Q, Crowley M, et al. A biophysical perspective on the cellulosome: new opportunities for biomass conversion. Curr Opin Biotechnol. 2008;19:218–227. | |
Krauss J, Zverlov V, Schwarz W. In vitro reconstruction of the complete Clostridium thermocellum cellulosome and synergistic activity on crystalline cellulose. Appl Environ Microbiol. 2012;78(12):4301–4307. | |
Bayer E, Belaich J-P, Shoham Y, Lamed R. The cellulosome: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. | |
Currie D, Herring C, Guss A, Olson D, Hogsett D, Lynd L. Functional heterologous expression of an engineered full length CipA from Clostridium thermocellum in Thermoanaerobacterium saccharolyticum. Biotechnol Biofuels. 2013;6:32–42. | |
Garvey M, Klose H, Fischer R, Lambertz C, Commandeur U. Cellulases for biomass degradation: comparing recombinant cellulose expression platforms. Trends Biotechnol. 2013;31(10):581–593. | |
Kovács K, Willson BJ, Schwarz K, et al. Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels. 2013;6:117–131. | |
Mazzoli R, Lambertz C, Pessione E. Engineering new metabolic capabilities in bacteria: lessons from recombinant cellulolytic strategies. Trends Biotechnol. 2012;30(2):111–119. | |
Wieczorek A, Martin V. Engineering the cell surface display of cohesions for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb Cell Fact. 2010;9:69–82. | |
Anderson TD, Robson SA, Jiang XW, et al. Assembly of minicellulosomes on the surface of Bacillus subtilis. Appl Environ Microbiol. 2011;77(14):4849–4858. | |
Levasseur A, Pagès S, Fierobe HP, et al. Design and production in Aspergillus niger of a chimeric protein associating a fungal feruloyl esterase and a clostridial dockerin domain. Appl Environ Microbiol. 2004;70:6984–6991. | |
Lilly M, Fierobe H-P, van Zyl W, Volschenk H. Heterologous expression of a Clostridium minicellulosome in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:1236–1249. | |
Tsai S-L, Oh J, Singh S, Chen R, Chen W. Functional assembly of minicellulosome on the Saccharomyces cerevisiae surface for cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2009;75:6087–6093. | |
Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol. 2010;76:1251–1260. | |
Kim S, Baek S-H, Lee K, Hahn J-S. Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb Cell Fact. 2013;12:14–21. | |
Vazana Y, Barak Y, Unger T, et al. A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnol Biofuels. 2013;6:182–200. | |
Moraïs S, Heyman A, Barak Y, et al. Enhanced cellulose degradation by nano-complexed enzymes: Synergism between a scaffold-linked exoglucanase and a free endoglucanase. J Biotechnol. 2010;147:205–211. | |
Mitsuzawa S, Kagawa H, Li Y, Chan S, Paavola C, Trent J. The rosettazyme: a synthetic cellulosome. J Biotechnol. 2009;143:139–144. | |
Gefen G, Anbar M, Morag E, Lamed R, Bayer E. Enhanced cellulose degradation by targeted integration of a cohesion-fused β-glucosidase into Clostridium thermocellum cellulosome. Proc Natl Acad Sci U S A. 2012;109(26):10298–10303. | |
Cunha E, Hatem C, Barrick D. Insertion of endocellulase catalytic domains into thermostable consensus ankyrin scaffolds: effects on stability and cellulolytic activity. Appl Environ Microbiol. 2013;79:6684–6696. | |
Tsai S, Park M, Chen W. Size-modulated synergy of cellulose clustering for enhanced cellulose hydrolysis. Biotechnol J. 2013;8:m257–m261. | |
Sun Q, Madan B, Tsai S, DeLisa M, Chen W. Creation of artificial cellulosomes on DNA scaffolds by zinc finger protein-guided assembly for efficient cellulose hydrolysis. Chem Commun. 2014;50:1423–1425. | |
Mazzoli R. Development of microorganisms for cellulose-biofuel consolidated bioprocessing: metabolic engineers’ tricks. Comput Struct Biotechnol J. 2012;3(4):1–9. | |
Hauser L, Land M, Larimer F. Clostridium Thermocellum ATCC 27405 Analysis Files; 2010. Available from: http://genome.ornl.gov/microbial/cthe. Accessed November 23, 2014. | |
Hemme CL, Mouttaki H, Lee YJ, et al. Sequencing of Multiple Clostridial Genomes related to biomass conversion and biofuel production. J Bacteriol. 2010;192(24):6494–6496. | |
Riederer A, Takasuka TE, Makino S, et al. Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose. Appl Environ Microbiol. 2011;77(4):1243–1253. | |
Raman B, McKeown C, Rodriguez M, Brown S, Mielenz J. Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation. BMC Microbiol. 2011;11:134–149. | |
Wei H, Fu Y, Magnusson L, et al. Comparison of transcriptional profiles of Clostridium thermocellum grown on cellobiose and pretreated yellow poplar using RNA-Seq. Front Microbiol. 2014;5(142):1–16. | |
Wilson CM, Rodriguez M Jr, Johnson CM, et al. Global transcriptome analysis of Clostridium thermocellum ATCC 27405 during growth on dilute acid pretreated Populus and switchgrass. Biotechnol Biofuels. 2013;6:179–198. | |
Rydzak T, McQueen PD, Krokhin OV, et al. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 2012;12:214–232. | |
Gold N, Martin V. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J Bacteriol. 2007;189:6787–6795. | |
Raman B, Pan C, Hurst GB, et al. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition; a quantitative proteomic analysis. PLoS One. 2009;4(4):5271–5284. | |
Roberts S, Gowen C, Brooks J, Fong S. Genome-scale metabolic analysis of Clostridium thermocellum for bioethanol production. BMC Syst Biol. 2010;4:31–48. | |
Tripathi SA, Olson DG, Argyros DA, et al. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbol. 2010;76:6591–6599. | |
Li Y, Tschaplinski TJ, Engle NL, et al. Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol Biofuels. 2012;5:2–15. | |
Yee KL, Rodriguez M Jr, Thompson OA, et al. Consolidated bioprocessing of transgenic switchgrass by an engineered and evolved Clostridium thermocellum strain. Biotechnol Biofuels. 2014;7:75–81. | |
Shao X, Raman B, Zhu M, et al. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl Microbiol Biotechnol. 2011;92:641–652. | |
Zhu X, Cui J, Feng Y, Fa Y, Zhang J, Cui Q. Metabolic adaption of ethanol-tolerant Clostridium thermocellum. PLoS One. 2013;8(7):70631–70640. | |
Brown SD, Guss AM, Karpinets TV, et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A. 2011;108:13752–13757. | |
Shaw AJ, Podkaminer KK, Desai SG, et al. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yields. Proc Natl Acad Sci U S A. 2008;105(37):13769–13774. | |
Ng T, Ben-Bassat A, Zeikus J. Ethanol production by thermophilic bacteria: fermentation of cellulosic substrates by cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1981;41(6):1337–1343. | |
Wiegel J, Ljungdahl L, Rawson J. Isolation from soil and properties of the extreme thermophile Clostridium thermohydrosulfuricum. J Bacteriol. 1979;138:800–810. | |
Xu L, Tschirner U. Improved ethanol production from various carbohydrates through anaerobic thermophilic co-culture. Bioresour Technol. 2011;102(21):10065–10071. | |
Cheng J, Zhu M. A novel anaerobic co-culture system for bio-hydrogen production from sugarcane bagasse. Bioresour Technol. 2013;144:623–631. | |
Linger J, Darzins A. Consolidated bioprocessing. In: Lee J, editor. Advanced Biofuels and Bioproducts. New York: Springer; 2013:267–280. | |
Higashide W, Li Y, Yang Y, Liao J. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl Environ Microbiol. 2011;77(8):2727–2733. | |
Li H, Cann A, Liao J. Biofuels: biomolecular engineering fundamentals and advances. Annu Rev Chem Biomol Eng. 2010;1(4):19–36. | |
Berezina O, Zakharova N, Yarotsky C, Zverlov V. Microbial producers of butanol. Appl Biochem Microbiol. 2012;48(7):625–638. | |
Yu E, Chan M, Saddler J. Butanol production from cellulosic substrates by sequential co-culture of Clostridium thermocellum and C. acetobutylicum. Biotechnol Lett. 1985;7(7):509–514. | |
Wen Z, Wu M, Lin Y, Yang L, Lin J, Cen P. A novel strategy for sequential co-culture of Clostridium thermocellum and Clostridium beijerinckii to produce solvents from alkali extracted corn cobs. Process Biochem. 2014;49:1941–1949. | |
Zuroff T, Xiques S, Curtis W. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels. 2013;6:59–71. | |
Zuroff T, Curtis W. Developing symbiotic consortia from lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93:1423–1435. | |
Bernstein HC, Kesaano M, Moll K, et al. Direct measurement and characterization of active photosynthesis zones inside wastewater remediating and biofuel producing microalgal biofilms. Bioresour Technol. 2014;156:206–215. | |
Kesaano M, Sisms R. Algal biofilm based technology for wastewater treatment. Algal Res. 2014;5:231–240. | |
Liu T, Wang J, Hu Q, et al. Attached cultivation technology of microalgae for efficient biomass feedstock production. Bioresour Technol. 2013;127:216–222. | |
Christenson L, Sims R. Rotating Algal biofilm reactor and spool harvester for wastewater treatment with biofuels by-products. Biotechnol Bioeng. 2012;109(7):1674–1684. | |
Wang Z-W, Chen S. Potential of biofilm-based biofuel production. Appl Microbiol Biotechnol. 2009;83:1–18. | |
Zuroff T, Gu W, Fore R, Leschine S, Curtis W. Insights into Clostridium phytofermentans biofilm formation: aggregation, micro-colony development and the role of extracellular DNA. Microbiology. 2014;160(6):1134–1143. | |
Dumitrache A, Wolfaardt G, Allen D, Liss S, Lynd L. Form and Function of Clostridium thermocellum biofilms. Appl Environ Microbiol. 2013;79(1):231–239. | |
Casolaro, K. Mascoma Announces Major Cellulosic Biofuel Technology Breakthrough; May 2009. Available from: http://www.mascoma.com/assets/Technology-AdvancesRelease-050709-FINAL.pdf. Archived Press. Accessed December 2, 2014. | |
Mascoma Corporation. Consolidated Bioprocessing (CBP) for High Efficiency Fermentation. Available from: http://www.mascoma.com/technology/consolidated-bioprocessing/. Accessed December 2, 2014. | |
Mascoma Corporation. Products Overview. Available from: http://www.mascoma.com/applications-products/products/. Accessed December 2, 2014. | |
Doran, K. Mascoma Awarded $80 Million from the DOE for Construction of Commercial-Scale Hardwood Cellulosic Ethanol Facility in Kinross [DOE press release]. Michigan; Dec 2011. Available from: http://www.mascoma.com/assets/Mascoma-_-DOE-Press-Release-FINAL.pdf. Accessed December 2, 2014. | |
QTEROS. Technology Overview. Available from: http://www.qteros.com/technology/overview/. Accessed December 2, 2014. | |
QTEROS. Qteros and UMass Amherst announce the issuance of a US Patent for production of ethanol from a unique microorganism, the Q Microbe™ [press release]; 2010. Available from: http://www.qteros.com/news/category/press/. Accessed December 2, 2014. | |
QTEROS. Simplified Conversion Process. Available from: http://www.qteros.com/technology/simplified-conversion-process/. Accessed December 2, 2014. | |
Kelly P. SunEthanol (Qteros) Wins 3rd DOE Grant to Make Ethanol from Non-food Plant Materials using its Q Microbe™; May 2008. Available from: http://www.qteros.com/news/press/11/. In Press 2009. Accessed December 2, 2014. | |
Durre P. Handbook on Clostridia. Boca Raton, FL: Taylor and Francis Group; 2005:109–112. | |
Guedon E, Desvaux M, Petitdemange H. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl Environ Microbiol. 2002;68:53–58. | |
Giallo J, Gaudin C, Belaich J. Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl Environ Microbiol. 1985;49:1216–1221. | |
Gardner J, Keating D. Requirement of the Type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Appl Environ Microbiol. 2010;76:5079–5087. | |
Dharmagadda V, Nokes S, Strobel H, Flythe M. Investigation of the metabolic inhibition observed in solid-substrate cultivation of Clostridium thermocellum on cellulose. Bioresour Technol. 2010;101:6039–6044. | |
Chinn M, Nokes S, Strobel H. Influence of moisture content and cultivation duration on Clostridium thermocellum 27405 end-product formation in solid substrate cultivation on Avicel. Bioresour Technol. 2008;99:2664–2671. | |
Tolonen A, Haas W, Chilaka A, Aach J, Gygi S, Church G. Proteome-wide systems analysis of a cellulosic biofuel producing microbe. Mol Syst Biol. 2011;7(1):461–473. | |
Jin M, Balan V, Gunawan C, Dale B. Consolidated Bioprocessing (CBP) Performance of Clostridium phytofermentans on AFEX-Treated Corn Stover for Ethanol Production. Biotechnol Bioeng. 2011;108:1290–1297. | |
Zambare V, Bhalla A, Muthukumarappan K, Sani R, Christopher L. Bioprocessing of agricultural residues to ethanol utilizing a cellulolytic extremophile. Extremophiles. 2011;15:611–618. | |
Okamoto K, Nitta Y, Maekawa N, Yanase H. Direct ethanol production from starch, wheat bran and rice straw by the white rot fungus Trametes hirsuta. Enzyme Microb Technol. 2011;48:273–277. | |
Lee J, Venditti R, Jameel H, Kenealy W. Detoxification of woody hydrolyzates with activated carbon for bioconversion to ethanol by the thermophilic anaerobic bacterium Thermoanaerobacterium saccharolyticum. Biomass Bioenergy. 2011;35:626–636. | |
Cripps RE, Eley K, Leak DJ, et al. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab Eng. 2009;11:398–408. | |
Cai Y, Lai C, Li S, et al. Disruption of lactate dehydrogenase through homologous recombination to improve bioethanol production in Thermoanaerobacterium aotearoense. Enzyme Microb Technol. 2011;48:155–161. | |
Yao S, Mikkelsen MJ. Metabolic engineering to improve ethanol production in Thermoanaerobacter mathranii. Appl Microbiol Biotechnol. 2010;88:199–208. | |
Lamed R, Lobos J, Su T. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microbiol. 1988;54(5):1216–1221. | |
Lv W, Yu Z. Isolation and characterization of two thermophilic cellulolytic strains of Clostridium thermocellum from compost sample. J Appl Microbiol. 2012;114:10001–10007. | |
Rydzak T, Levin D, Cicek N, Sparling R. End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol. 2011;92:199–209. | |
Liu Y, Yu P, Song X, Qu Y. Hydrogen production from cellulose by co-culture of Clostridium thermocellum JN4 and Thermoanaerobacterium thermosaccharolyticum GD17. Int J Hydrogen Energy. 2008;33:2927–2933. | |
Freier D, Mothershed C, Wiegel J. Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol. 1988;54(1):204–211. | |
Geng A, He Y, Qian C, Yan X, Zhou Z. Effects of key factors on hydrogen production from cellulose in a co-culture of Clostridium thermocellum and Clostridium thermopalmarium. Bioresour Technol. 2010;101:4029–4033. | |
Balusu R, Paduru R, Kuravi S, Seenaya G, Reddy G. Optimization of critical medium components using response surface methodology for ethanol production from cellulosic biomass by Clostridium thermocellum SS19. Process Biochem. 2005;40:3025–4030. | |
Rani K, Swamy M, Seenaya G. Production of ethanol from various pure and natural cellulosic biomass by Clostridium thermocellum strains SS21 and SS22. Process Biochem. 1998;33:435–440. | |
Nigro O, Culley A, Steward G. Complete genome sequence of bacteriophage VvAW1, which infects Vibrio vulnificus. Stand Genomic Sci. 2012;6:415–426. | |
Tamaru Y, Karita S, Ibrahim A, Chan H, Doi R. A large gene cluster for the Clostridium cellulovorans cellulosome. J Bacteriol. 2000;182:5906–5910. | |
Ding S, Bayer E, Steiner D, Shoham Y, Lamed R. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J Bacteriol. 1999;181:6720–6729. | |
Bayer E, Ding S, Shoham Y, Lamed R. New perspectives in the structure of cellulosome-related domains from different species. In: Ohimya K, Hayashi K, editors. Genetics, Biochemistry and Ecology of Lignocellulose Degradation. Tokyo: Uni Publishers Co; 1999:428–436. | |
Lamed R, Morag E, Moryosef O, Bayer E. Cellulosome-like entities in Bacteroides cellulosolvens. Curr Microbiol. 1991;22:27–34. | |
Ding S, Bayer E, Steiner D, Shoham Y, Lamed R. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J Bacteriol. 2000;182:4915–4925. | |
Ponpium P, Ratanakhanokchai K, Kyu K. Isolation and properties of a cellulosome-type multienzyme complex of the thermophilic Bacteroides sp. strain P-1. Enzyme Microb Technol. 2000;26:459–465. | |
Sleat R, Mah R, Robinson R. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984;48:88–93. | |
Shoseyov O, Doi R. Essential 170 kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc Natl Acad Sci U S A. 1990;87:2192–2195. | |
Bélaich J, Tardif C, Bélaich A, Gaudin C. The cellulolytic system of Clostridium cellulolyticum. J Biotechnol. 1997;57:3–14. | |
Pagès S, Bélaïch A, Bélaïch JP, et al. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins. 1997;29:517–527. | |
Pagès S, Bélaïch A, Fierobe H, Tardif C, Gaudin C, Bélaïch J. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181:1801–1810. | |
Kakiuchi M, Isui A, Suzuki K, et al. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180: 4303–4308. | |
Sakka K, Karita S, Ohmiya K, Shimada K. Xylanases of Clostridium stercorarium and Clostridium thermocellum isolated in Japan. In: Shimada K, Hoshino S, Ohmiya K, Sakka K, Kobayashi Y, Karita S, editors. Genetics, Biochemistry and Ecology of Lignocellulose Degradation. Tokyo: Uni Publishers; 1994:32–38. | |
Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. | |
Gerngross U, Romaniec M, Kobayashi T, Huskisson N, Demain A. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8:325–334. | |
Ohara H, Karita S, Kimura T, Sakka K, Ohmiya K. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci Biotechnol Biochem. 2000;64:254–260. | |
Kirby J, Martin J, Daniel A, Flint H. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett. 1997;149:213–219. | |
Ding SY, Rincon MT, Lamed R, et al. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J Bacteriol. 2001;183: 1945–1953. | |
Ljungdahl L. The cellulose/hemicellulose system of the anaerobic fungus Orpinomyces PC-2 and aspects of its applied use. Ann N Y Acad Sci. 2008;1125:308–321. | |
Steenbakkers PJ, Li XL, Ximenes EA, et al. Noncatalytic docking domains of cellulosomes of anaerobic fungi. J Bacteriol. 2001; 183(18):5325–5333. | |
Li X, Chen H, Ljungdahl L. Two cellulases, CelA and CelC, from the polycentric anaerobic fungus Orpinomyces strain PC-2 contain N-terminal docking domains for a cellulase-hemicellulase complex. Appl Environ Microbiol. 1997;63:4721–4728. | |
Fanutti C, Ponyi T, Black G, Hazelwood G, Gilbert H. The conserved noncatalytic 40-residue sequences in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J Biol Chem. 1995;270(40):29314–29322. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


