Back to Journals » Drug Design, Development and Therapy » Volume 8
Clinicopathological significance and potential drug target of RUNX3 in breast cancer
Authors Yu Y, Chen C, Kong F, Zhang W
Received 26 July 2014
Accepted for publication 21 August 2014
Published 5 December 2014 Volume 2014:8 Pages 2423—2430
DOI https://doi.org/10.2147/DDDT.S71815
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Shu-Feng Zhou
Ying-Ying Yu,1 Chao Chen,2 Fan-fei Kong,2 Wei Zhang1
1Obstetrics and Gynecology Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Gynecology, Shanghai First Maternity and Infant Hospital Affiliated to TongJi University, Shanghai, People’s Republic of China
Background: Previous reports indicate that RUNX3 is a tumor suppressor in several types of human tumors, including breast cancer (BC). However, the correlation between RUNX3 hypermethylation and the incidence of BC remains unclear. In this study, we conducted a systematic review and meta-analysis aiming to comprehensively assess the potential role of RUNX3 hypermethylation in the pathogenesis of BC.
Methods: A detailed literature search was made to identify studies for related research publications. Methodological quality of the studies was evaluated. Analysis of pooled data was performed. Odds ratio (OR) was calculated and summarized respectively.
Results: Final analysis of 565 BC patients from eleven eligible studies was performed. The results showed that RUNX3 hypermethylation was significantly higher in BC than in normal breast tissue, the pooled OR from nine studies including 339 BC and 248 normal breast tissue (OR =24.12, 95% confidence interval [CI] =13.50–43.11, Z=10.75, P<0.00001). Further analysis also showed significantly increased OR of RUNX3 hypermethylation in estrogen receptor (ER)-positive than in ER-negative BC patients (OR =5.67, 95% CI =2.69–11.95, Z=4.57, P<0.00001). In addition, RUNX3 messenger RNA (mRNA) high expression was found to be correlated to better overall survival in 3,455 cases of BC patients that were followed up for 20 years (hazard ratio [HR] 0.79, P=8.8×10-5). Interestingly, RUNX3 mRNA overexpression was found to be correlated to better overall survival in only 668 cases of ER-negative patients (HR 0.72, P=0.01), but not in 1,767 cases of ER-positive patients (HR 0.87, P=0.13).
Conclusion: The results of this meta-analysis suggest that RUNX3 hypermethylation may be implicated in the pathogenesis of BC. Detection of RUNX3 mRNA may be a helpful and valuable biomarker for diagnosis of BC, especially in ER-negative BC. We also discussed the significance of RUNX3 as a potential drug target.
Keywords: methylation, tumor suppressor gene, meta-analysis, odds ratio
Introduction
Breast cancer (BC), with approximately 232,340 new cases every year, is the most common malignancy among women and is also the leading cancer-related death for women worldwide.1 Although early diagnostic methods, surgical techniques, and molecular target therapy have undergone considerable advancements, the incidence of BC is still increasing, and the prognosis of BC patients remains discouraging due to the high postoperative recurrence rate and metastasis.2,3 Therefore, investigations on the mechanism of incidence and progression of BC are still needed and will help to select patients with higher chances of BC recurrence and provide better prognostic prediction and individualized treatments.
The runt-domain related (RUNX) family genes are essential regulator genes of cell fate in development and regulation of p53-dependent DNA damage response and/or tumorigenesis.4–6 RUNX family genes are composed of RUNX1, RUNX2, and RUNX3. Among the three members, RUNX3 gene plays a critical role in the regulation of cell proliferation, apoptosis, angiogenesis, as well as cell adhesion and invasion.7,8 RUNX3 gene is considered as a tumor suppressor gene involved in the TGF-β signaling pathway because it is localized in chromosome 1p36, a region that exhibits frequent loss of heterozygosity events in breast, colon, gastric, and ovarian cancers.9 RUNX3 is one of principal responders of the p14(ARF)-MDM2 cell surveillance pathway, and it is able to prevent pathologic consequences of abnormal oncogene activation.10 The molecular and biological functions of RUNX3 have been intensively studied in several types of tumors through upregulation of inducing cell cycle arrest, apoptosis, and downregulation of cyclin D1 expression.11–15 Downregulation of RUNX3 protein by promoter methylation (hypermethylation) has been found to play an important role in epithelial tumorigenesis and epithelial-mesenchymal transition of several malignancies including BC.11,15–21 RUNX3 also inhibits the estrogen-dependent proliferation, transformation, and the tumorigenicity of BC cells in severe combined immunodeficiency mice.22 Of female RUNX3 (+/−) mice, 20% spontaneously developed ductal carcinoma at an average age of 14.5 months, indicating that RUNX3 acts as a novel tumor suppressor in BC.22
Although previous studies have indicated that inactivation of the RUNX3 gene is mainly induced by its promoter hypermethylation and is one of the important epigenetic alterations in BC, the reported rates of RUNX3 hypermethylation in BC are remarkably diverse. Moreover, whether or not RUNX3 gene hypermethylation is associated with the incidence of BC remains unclear. The various results of these studies underpin the need for assessing the evidence of the relationship between RUNX3 inactivation and causes of BC. In the current study, we performed a systematic review and meta-analysis to quantitatively evaluate the effects of RUNX3 hypermethylation on the incidence of BC. In addition, we evaluated RUNX3 messenger RNA (mRNA) as a prognostic marker in BC patients and discussed RUNX3 as a potential drug target.
Material and methods
Search strategy and selection criteria
We searched PubMed, Embase, and ISI web of knowledge to identify studies from January 1, 2000 to July, 2014 using the search terms: “breast” and “cancer OR tumor OR neoplasm OR carcinoma”, “methylation”, and “RUNX3”. We also manually searched the reference lists of the retrieved articles and reviews for additional articles.
After excluding the nonrelevant and/or redundant publications from the different databases, the remaining papers were evaluated in the full-text version for inclusion and exclusion criteria and for relevant articles in the reference lists. All searched data were retrieved. Authors’ bibliographies and references of selected studies were also searched for additional relevant studies. The most complete study was chosen to avoid duplication if same patient populations were reported in different publications.
Criteria that an eligible study had to meet were as follows: 1) RUNX3 methylation evaluated in the primary BC tissues, 2) research revealed the relationship between RUNX3 methylation and BC incidence, and 3) studies provided sufficient information to estimate an odds ratio (OR) and 95% confidence interval (CI). The exclusion criteria included the following: 1) letters, reviews, case reports, conference abstracts, editorials, and expert opinion; and 2) all publications regarding in vitro/ex vivo studies, cell lines, and human xenografts were also excluded.
Data extraction and methodological assessment
We reviewed and extracted data from eligible studies. Disagreements were resolved by discussion and consensus. The following information were recorded for each study: the first author name, year of publication, authors’ country, sample source, number of cases, clinicopathological parameters, methylation detection method, methylation rate and/or expression, and follow up. Data for study characteristics were summarized in a table format. Heterogeneity of investigation was evaluated to determine whether the data of the various studies could be analyzed for a meta-analysis.
For the methodological evaluation of the studies, we read through each publication independently and assessed and scored the publications according to REMARK guidelines and ELCWP quality scale.23,24 We shared the quality scores and compared them. Then, we reached a consensus value for each item.
Patient survival analysis
An online database25 was used to assess the relevance of RUNX3 mRNA expression to relapse-free survival. The database was established using gene expression data and survival information of 3,455 BC patients downloaded from Gene Expression Omnibus. Briefly, RUNX3 gene was entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=breast) to obtain Kaplan–Meier survival plots where the number-at-risk is indicated below the main plot. Hazard ratio (and 95% CIs) and log rank P were calculated and displayed on the webpage.
Statistical analysis
Analysis was conducted using the STATA 12.0 (StataCorp LP, College Station, TX, USA) and Review Manager 5.2 (Cochrane Collaboration, Oxford, UK). The pooled frequency of RUNX3 hypermethylation and 95% CIs were estimated. The frequency of RUNX3 hypermethylation was compared in BC and normal tissues. Heterogeneity among studies was evaluated with Cochran’s Q test26 and the I2 statistic.27,28 When heterogeneity was not an issue (I2 values <50%), a fixed effect model was used to calculate parameters. If there was substantial heterogeneity (I2 values ≥50%), a random-effects model was used to pool data and attempt to identify potential sources of heterogeneity based on subgroup analyses. The pooled OR was estimated for RUNX3 hypermethylation in BC and in normal breast tissues. P-values tailed less than 0.05 were considered statistically significant.
Publication bias was assessed by using a method reported by Egger et al.29 We also explored reasons for statistical heterogeneity using meta-regression, subgroup analysis, and sensitivity analysis. The analysis of meta-regression and publication bias was performed using STATA version 10.0.
Results
Identification of relevant studies
Fifty-five publications were identified by the search method as described above. Forty-four of those were excluded due to laboratory studies, non-original articles (review), or studies irrelevant to the current analysis. Eventually, there were eleven studies included in the final meta-analysis, as shown in Figure 1.
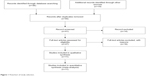  | Figure 1 Flowchart of study selection. |
Study characteristics
Eleven studies published from 2005 to 2013 were eligible for meta-analysis. A total of 565 BC patients from the People’s Republic of China, South Korea, Japan, Singapore, and USA were enrolled. Their basic characteristics were summarized in Table 1.
The correlation of RUNX3 hypermethylation with carcinogenesis
Comparison of RUNX3 hypermethylation in BC tissue and normal breast tissue
First, we determined that RUNX3 hypermethylation was significantly higher in BC than in normal breast tissues. There was no evidence of heterogeneity across the studies (P for heterogeneity =0.42; I2 =2%). The pooled OR from nine studies including 339 BC and 248 normal breast tissue is shown in Figure 2 (OR =24.12, 95% CI =13.50–43.11, Z=10.75, P<0.00001), indicating that RUNX3 inactivation through hypermethylation plays an important role in the pathogenesis of BC.
Correlation of RUNX3 hypermethylation with estrogen receptor status in BC
Then, we determined whether or not RUNX3 hypermethylation rate in BC was correlated to estrogen receptor (ER) status in BC patients. The pooled OR from three studies including 139 ER-positive BC and 101 ER-negative BC is shown in Figure 3 (OR =5.67, 95% CI =2.69–11.95, Z=4.57, P<0.00001), indicating that RUNX3 hypermethylation was significantly higher in ER-positive BC than in ER-negative BC.
Sensitivity analyses and publication bias
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled OR was not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (Figure 4A and B), suggesting there were no publication biases in the meta-analysis.
Impact of RUNX3 mRNA expression on prognosis of BC
The clinical relevance of RUNX3 was further corroborated in a patient survival analysis using an online database containing the expression of 22,277 genes and 20-year survival information of 3,455 BC patients.25 RUNX3 mRNA high expression was found to be correlated to better overall survival (OS) for all BC patients followed for 20 years (Figure 5A; hazard ratio [HR] 0.79, P=8.8×10−5). In addition, RUNX3 mRNA high expression was not found to be correlated to better OS in ER-positive patients (Figure 5B; HR 0.87, P=0.13) but only in ER-negative patients (Figure 5C; HR 0.72, P=0.01).
Discussion
Inactivation of RUNX3 by promoter hypermethylation plays an important role during normal development and tumorigenesis in several types of tumors including BC.11,30–40 To date, there have been some studies describing the precise expression and methylation status of RUNX3 in BC; however, the roles of RUNX3 hypermethylation in BC and its correlation with carcinogenesis have not been thoroughly investigated. Analysis of the pooled data showed that BC had a higher RUNX3 hypermethylation than normal breast tissues. RUNX3 hypermethylation was also significantly higher in ER-positive BC than in ER-negative BC. The results from the current study indicated that the hypermethylation rate of RUNX3 gene promoter in BC was strongly correlated to BC incidence. Since changes in RUNX3 promoter hypermethylation are reversible, drug treatment through demethylation may be useful to delay carcinogenesis and progression. Trichostatin A and/or 5-Aza-CdR (5-aza-2′-deoxycytidine) are capable of inhibiting BC cell proliferation and inducing apoptosis by eliminating the methylation status of RUNX3 promoter and restoring its expression.41,42 Reintroduction of RUNX3 into UNX3-deficient BC cells suppresses cancer cell proliferation and their tumorigenic potential.22,42,43 Therefore, restoring RUNX3 activation by specific small molecules or inhibitors to block RUNX3 hypermethylation might constitute a novel therapeutic strategy for the treatment of BC. This approach brings a new direction and hope for cancer treatment through epigenetic modulation and/or gene-targeted therapy.
Epigenetic alterations, particularly aberrant DNA methylation, one of the best-characterized epigenetic modifications, contribute to tumor initiation and progression.44,45 RUNX3 inhibits the oncogenic Wnt signaling pathway via the formation of a complex with the TCF4-β-catenin complex and hampering it from binding to target genes such as c-myc and cyclin D1.18,30 RUNX3 interacts with SMAD3/SMAD4 to activate TGF-β-dependent proliferation inhibition and apoptosis by the activation of p21 and Bim. In RUNX3 (−/−) p53 (−/−) murine gastric epithelial (GIF-14) cells, concurrent activation of an EGFR/Ras gene expression signature was observed during TGF-β-induced epithelial-mesenchymal transition in GIF-14 cells.19 RUNX3 also destabilizes HIF-1α protein by promoting the proline hydroxylation of HIF-1α through binding to HIF-1α/PHD2, thus RUNX3 could be an inhibitor of HIF-1α and of hypoxia-mediated angiogenesis.46 RUNX3 acts as a novel co-activator for p53 through regulating its DNA damage-induced phosphorylation at Ser-15 and mediates tumor suppression.47 In addition, RUNX3 could also inhibit ERα-dependent transactivation by reducing the stability of ERα in BC.22 Therefore, RUNX3 can be considered as a tumor suppressor, and its inactivation could contribute to tumor initiation and progression. Based on this meta-analysis, we may conclude that RUNX3 hypermethylation in BC tends to indicate higher incidence of BC. We further determined the clinical relevance of RUNX3 mRNA expression in a patient survival analysis using an online database containing the expression of 22,277 genes and 20-year survival information of 3,455 BC patients.25 RUNX3 mRNA high expression was found to be correlated to better OS for all BC patients (HR 0.79, P=8.8×10−5). Interestingly, RUNX3 mRNA high expression was not found to be correlated to better OS in ER-positive patients (HR 0.87, P=0.13) but only in ER-negative patients (HR 0.72, P=0.01). RUNX3 mRNA low expression might be due to the RUNX3 hypermethylation in BC patients. Since ER-negative BC patients lack the prognostic marker and drug target, RUNX3 will be more important for future study in ER-negative BC.
Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found. This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders could not be completely excluded because all of the included studies were observational. Second, the searching strategy was restricted to articles published in English and Chinese. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. The correlation of RUNX3 hypermethylation with ER status in BC was based on the pooled OR from only three studies including 139 ER-positive BC and 101 ER-negative BC; the conclusion will need further validation in future study. In addition, most selected publications are from Asia; none are from Europe. Hence, cautions should be taken when our findings are interpreted among the general populations.
In conclusion, our meta-analysis showed RUNX3 may play an important role in BC incidence, and RUNX3 hypermethylation is strongly correlated with ER status. In addition, RUNX3 mRNA high expression was found to be correlated to better OS for all BC patients, especially in ER-negative BC patients. Further large-scale studies, especially multicenter and well-matched cohort research, will provide more insight into the role of RUNX3 in the carcinogenesis and clinical implementation of BC patients.
Disclosure
The authors have no financial involvement with any organization or entity with a financial interest in the subject matter or materials discussed in the manuscript.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. | ||
Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274:113–126. | ||
Coleman RE, Gregory W, Marshall H, Wilson C, Holen I. The metastatic microenvironment of breast cancer: clinical implications. Breast. 2013;22 Suppl 2:S50–S56. | ||
Ozaki T, Nakagawara A, Nagase H. RUNX Family participates in the regulation of p53-dependent DNA damage response. Int J Genomics. 2013;2013:271347. | ||
Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. | ||
Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. | ||
Subramaniam MM, Chan JY, Yeoh KG, Quek T, Ito K, Salto-Tellez M. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta. 2009;1796:315–331. | ||
Lund AH, van Lohuizen M. RUNX: a trilogy of cancer genes. Cancer Cell. 2002;1:213–215. | ||
Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. | ||
Chi XZ, Kim J, Lee YH, et al. Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res. 2009;69:8111–8119. | ||
Shiraha H, Nishina S, Yamamoto K. Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J Cell Biochem. 2011;112:745–749. | ||
Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. | ||
Chi XZ, Yang JO, Lee KY, et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol Cell Biol. 2005;25:8097–8107. | ||
Wei D, Gong W, Oh SC, et al. Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res. 2005;65:4809–4816. | ||
Chen LF. Tumor suppressor function of RUNX3 in breast cancer. J Cell Biochem. 2012;113:1470–1477. | ||
Tanaka S, Shiraha H, Nakanishi Y, et al. Runt-related transcription factor 3 reverses epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Cancer. 2012;131:2537–2546. | ||
Li J, Jiang X. Loss of runt-related transcription factor 3 expression associated with human hepatocellular carcinoma progression and prognosis. Asian Pac J Cancer Prev. 2011;12:2285–2290. | ||
Voon DC, Wang H, Koo JK, et al. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells. 2012;30:2088–2099. | ||
Voon DC, Wang H, Koo JK, et al. EMT-induced stemness and tumorigenicity are fueled by the EGFR/Ras pathway. PloS One. 2013;8:e70427. | ||
Liu Z, Chen L, Zhang X, et al. RUNX3 regulates vimentin expression via miR-30a during epithelial-mesenchymal transition in gastric cancer cells. J Cell Mol Med. 2014;18:610–623. | ||
Subramaniam MM, Chan JY, Omar MF, et al. Lack of RUNX3 inactivation in columnar cell lesions of breast. Histopathology. 2010;57:555–563. | ||
Huang B, Qu Z, Ong CW, et al. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor alpha. Oncogene. 2012;31:527–534. | ||
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180–1184. | ||
Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18:705–719. | ||
Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. | ||
DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15:1237–1248; discussion 49–52. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. | ||
Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29:2605–2615. | ||
Lee JH, Pyon JK, Kim DW, et al. Expression of RUNX3 in skin cancers. Clin Exp Dermatol. 2011;36:769–774. | ||
Lee JM, Shin JO, Cho KW, et al. Runx3 is a crucial regulator of alveolar differentiation and lung tumorigenesis in mice. Differentiation. 2011;81:261–268. | ||
Lee YM. Control of RUNX3 by histone methyltransferases. J Cell Biochem. 2011;112:394–400. | ||
Levanon D, Brenner O, Otto F, Groner Y. Runx3 knockouts and stomach cancer. EMBO Rep. 2003;4:560–564. | ||
Lee YS, Lee JW, Jang JW, et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell. 2013;24:603–616. | ||
Araki K, Osaki M, Nagahama Y, et al. Expression of RUNX3 protein in human lung adenocarcinoma: implications for tumor progression and prognosis. Cancer Sci. 2005;96:227–231. | ||
Subramaniam MM, Chan JY, Soong R, et al. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat. 2009;113:113–121. | ||
Hwang KT, Han W, Bae JY, et al. Downregulation of the RUNX3 gene by promoter hypermethylation and hemizygous deletion in breast cancer. J Korean Med Sci. 2007;22 Suppl:S24–S31. | ||
Mu WP, Wang J, Niu Q, Shi N, Lian HF. Clinical significance and association of RUNX3 hypermethylation frequency with colorectal cancer: a meta-analysis. Onco Targets Ther. 2014;7:1237–1245. | ||
Yang Y, Ye Z, Zou Z, Xiao G, Luo G, Yang H. Clinicopathological significance of RUNX3 gene hypermethylation in hepatocellular carcinoma. Tumour Biol. 2014;19:19. | ||
Kang HF, Dai ZJ, Bai HP, et al. RUNX3 gene promoter demethylation by 5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line. Onco Targets Ther. 2013;6:411–417. | ||
Lau QC, Raja E, Salto-Tellez M, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–6520. | ||
Chen W, Salto-Tellez M, Palanisamy N, et al. Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes Chromosomes Cancer. 2007;46:288–301. | ||
Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14:15029–15058. | ||
Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–316. | ||
Lee SH, Bae SC, Kim KW, Lee YM. RUNX3 inhibits hypoxia-inducible factor-1α protein stability by interacting with prolyl hydroxylases in gastric cancer cells. Oncogene. 2014;33(11):1458–1467. | ||
Yamada C, Ozaki T, Ando K, et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J Biol Chem. 2010;285:16693–16703. | ||
Li DX, Wu H, Gao JZ, Fang ZX. Relationship between methylation status of Runx3 gene promoter and Runx3 protein expression in patients with breast cancer. China Maternal and Child Health. 2013;28:3650–3653. | ||
Park SY, Kwon HJ, Choi Y, et al. Distinct patterns of promoter CpG island methylation of breast cancer subtypes are associated with stem cell phenotypes. Mod Pathol. 2012;25:185–196. | ||
Qiao L, Yang ZY, Liu W. Clinical significance of detecting plasma Runx3 gene promoter methylation in early diagnosis breast cancer. Mod Lab Med. 2012;27:35–37. | ||
Park SY, Kwon HJ, Lee HE, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458:73–84. | ||
Du JR, Lou G, Jiang Y. Relationship between the methylation of RUNX3 promoter and the clinical pathological characters and prognostic factors of breast invasive ductal carcinoma. Journal of Harbin Medical University. 2008;1:019. | ||
Tian SW, Jia SC, Chen J. Analysis of the status of Runx3 gene promoter methylation in breast cancer and its relationship with pathological features. Chin J Breast Dis. 2010;4:50–53. | ||
Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244–251. | ||
Suzuki M, Shigematsu H, Shames DS, et al. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93:1029–1037. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





