Back to Journals » International Journal of Nanomedicine » Volume 14
Capture and biological release of circulating tumor cells in pancreatic cancer based on peptide-functionalized silicon nanowire substrate
Authors Shen Q , Yang H, Peng C, Zhu H , Mei J, Huang S, Chen B, Liu J, Wu W, Cao S
Received 17 September 2018
Accepted for publication 4 December 2018
Published 24 December 2018 Volume 2019:14 Pages 205—214
DOI https://doi.org/10.2147/IJN.S187892
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Qinglin Shen,1,2,* Haitao Yang,3,* Caixia Peng,4,5,* Han Zhu,6 Jia Mei,1 Shan Huang,1 Bin Chen,7 Jue Liu,8 Wenbo Wu,3 Shaokui Cao3
1Department of Oncology, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; 2Cancer Center, Renmin Hospital, Wuhan University, Wuhan, China; 3School of Materials Science and Engineering, Zhengzhou University, Zhengzhou, China; 4Key Laboratory for Molecular Diagnosis of Hubei Province, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; 5Central Laboratory, The Central Hospital of Wuhanper, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China; 6Key Laboratory of Synthetic and Biological Colloids, Ministry of Education, School of Chemical and Material Engineering, Jiangnan University, Wuxi, China; 7Central Laboratory, Renmin Hospital, Wuhan University, Wuhan, China; 8Department of Pharmacy, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
*These authors contributed equally to this work
Background: Efficient and precise circulating tumor cells’ (CTCs) capture and release with minimal effect on cell viability for CTCs’ analysis are general requirements of CTCs’ detection device in clinical application. However, these two essential factors are difficult to be achieved simultaneously.
Methods: In order to reach the aforementioned goal, we integrated multiple strategies and technologies of staggered herringbone structure, nanowires’ substrate, peptides, enzymatic release, specific cell staining, and gene sequencing into microfluidic device and the sandwich structure peptide-silicon nanowires’ substrate was termed as Pe-SiNWS.
Results: The Pe-SiNWS demonstrated excellent capture efficiency (95.6%) and high release efficiency (92.6%). The good purity (28.5%) and cell viability (93.5%) of CTCs could be obtained through specific capture and biological release by using Pe-SiNWS. The good purity of CTCs facilitated precise and quick biological analysis, and five types of KRAS mutation were detected in 16 pancreatic cancer patients but not in healthy donors.
Conclusion: The results proved that the effective capture, minor damage release, and precise analysis of CTCs could be realized simultaneously by our novel strategy. The successful clinical application indicated that our work was anticipated to open up new opportunities for the design of CTC microfluidic device.
Keywords: circulating tumor cells, Pe-SiNWS, biological release, pancreatic cancers
Introduction
Pancreatic cancer typically has a very poor prognosis due to its features of rapid progression and deterioration, causing hundreds of thousands of deaths every year.1,2 More importantly, it usually has no symptoms in the disease’s early stages.2 So early diagnosis of pancreatic cancer is particularly important. As we all known, circulating tumor cells (CTCs) are disseminated cancer cells from primary or metastatic tumors that participate in blood circulation. CTCs carry significant information of tumors, which make their effective capture and analysis very critical to investigate cancer progression and metastasis.3 Therefore, early CTC detection of pancreatic cancer should be significant for clinical utility. However, CTCs existed in blood were really rare, making their detection a very challenging task. To address this issue, a variety of methods have been developed.4 Initially, immunomagnetic separation was mostly used to isolate CTCs, but its low capture efficiency and tedious steps severely limit its practical application.5,6 Recently, microfluidic technologies were developed to capture CTCs, which attracted more and more attention due to its unique advantages including automatic operation, high throughput, and superior sensitivity.6 In addition, by integrating other separation techniques, such as immunoaffinity purification,7 nanostructured surface,8 and size-based filtration system,9 into microfluidic devices, the efficiency of CTCs capture and analysis was further improved. Nevertheless, none of these methods fully serves the purpose for biological or clinical utility. Generally speaking, immunoaffinity purification often suffers from the unsatisfactory recovery of intact CTCs,10 while nanostructured surface6,8 and size-based filtration system10 lack specificity. New technologies are thus desired to address these issues in this field.
During the past two decades, scientists have developed a great quantity technology for capturing, releasing, and analyzing CTCs based on different principles. Mehmet Toner’s group combined deterministic lateral displacement, inertial focusing, and magnetic deflection parts together with microfluidic device that could capture both epithelial cell adhesion molecule (EpCAM, one of the common surface markers for CTCs)-positive and -negative CTCs by using two separation techniques.11 Subsequently, Moon et al12 combined multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP)-based separation method with microfluidic device, in which MOFF provided high-throughput filtration of blood cells, while DEP technique realized more precise capture. Rohani et al and Shim et al developed CTCs’ isolation and release method based on the continuous-flow DEP.13,14 Many other researchers also developed CTC capture and collection devices, which integrated different release methods (eg, trypsin-based assay,15 aptamer-based separation,16 temperature response polymer-based assay,17 and laser capture microdissection (LCM) technology18 into nanowire substrates). Despite the notable success of current microfluidic devices in the detection of CTCs, most of them are still unsatisfactory for actual application, since the release and analysis are equally important but less considered as compared to capture in clinical application.5,6,10,19 Thus, it is significant to explore new technologies and devices for actual application of CTCs’ detection.
Combining complementary separation techniques into microfluidic devices seemed to be a strategy to address these issues. In this contribution, we adopted and developed a new device for the actual application of CTCs’ detection, considering its good adaptability for integrating different techniques.13–16,20 In our design, two strategies are used to improve the capture efficiency. To enhance the interactions between cells and channel substrate and further improve CTC capture efficiency, unique staggered herringbone structure and nanowire were adopted in microfluidic device first (Figure 1A).21,22 On the other hand, we chose a polypeptide CKAAKN with specific affinity for pancreatic cancer cells.23 We hypothesize that the combination of EpCAM and specific peptides CKAAKN can enhance the capture efficiency of CTCs in pancreatic cancer patients, because the CKAAKN can capture the EpCAM-negative CTCs and specifically bind CTCs of pancreatic cancer. With regard to the release of CTCs, the peptide digestion is one of the biological methods, which have advantages in simple operation, minor damage and efficiently release, and minimal effect on cell viability compared to physical method13 and chemical method.24,25 The biological method is very important for later CTC analysis. In this work, for the first time, novel biological method of enzymatic release was adopted and explored, considering the enzyme specificity.
With above design, as shown in Figure 1, Pe-SiNWS was established, in which staggered herringbone structure was adopted for the fabrication of microfluidic device, and nanowires were integrated onto channel substrate (Figure 1A). Meanwhile, the strategies of peptide and enzymatic release were also adopted (Figure 1B). As a consequence, this Pe-SiNWS realized effective CTC capture (95.6%). After enzymatic releasing, obtained CTC could also achieve good purity (28.5%) and cell viability (93.5%). By using this device, CTCs can be collected from blood samples of patients with pancreatic cancer. Through gene sequencing, the KRAS mutation in pancreatic cancer can be detected quickly. These results prove that the effective capture, minor damage release, and analysis of CTCs can be realized simultaneously by our new strategy.
Materials and methods
Materials
Single Crystal Silicon Wafer was purchased from Western Minmetals (SC) Corporation. Sulfuric acid (98%), hydrogen peroxide (30%), silver nitrate (>99.8%), ammonium fluoride (≥99.99%), ethanol (>99.5%), and (3-aminopropyl)triethoxysilane (APTES, 99%) were purchased from Sigma-Aldrich. EZ-Link™ NHS-PEG4-Biotin was obtained from Thermo Fisher Scientific. Pancreatic cancer cell lines (BxPC3, Panc-1, and AsPC3) were purchased from American Type Culture Collection (ATCC; Rockville, MD, USA). White blood cells (WBCs) were collected from healthy donors. RPMI cell culture medium was obtained from Thermo Fisher Scientific. Custom peptides were ordered from Chinese Peptide Company (Hangzhou, China). Peptide digestion reagents (including ArgC, AspN, chymotrypsin, GluC, LysargiNase, LysC, LysN, pepsin, and trypsin) were obtained from Wako Chemicals (Richmond, VA, USA).
Methods to prepare silicon nanowires
Lithographically patterned silicon nanowires were prepared by a standard photolithography and a chemical wet etching process. First, polymethyl methacrylate (PMMA) A8 photoresist was spin coated on a silicon wafer (0.5 mm thick). After the exposure of UV light, the silicon wafer was kept in etching the solution of NH4F/AgNO3. Under the assistance of Ag nanoparticle deposition, there formed a template on the surface of the silicon wafer. Then, H2O2/NH4F was used to further etch the template for 10 min and was cleaned by H2SO4/H2O2 (70/30, v/v), deionized water, and absolute ethanol. After being dried with nitrogen, the patterned silicon nanowires were obtained.
Preparation of chemical modified and biological functioned silicon nanowires
First, photolithography and wet etching were used to introduce vertically aligned silicon nanowires onto a silicon wafer. Then, the washed and dried silicon wafer was soaked in a freshly prepared toluene solution of 3-aminopropyl triethoxysilane (APTES, 1.0 wt%) for 2 h. Afterward, the silicon wafer (surface-bonded with mercaptopropylmethoxysilane; MPTMS) was taken out and washed with absolute ethanol and dried with nitrogen. The modified silicon wafer with MPTMS was then soaked in a PBS solution (50 ng/mL, 200 mL) of NHS-PEGn-biotin (n=1,000) for 2 h. Then, the silicon wafer was taken out, washed with absolute ethanol, dried with nitrogen, and stored under dry environment at 4°C.
Prior to the blood sample test, the silicon wafer was washed with PBS and then incubated with 200 μL of streptavidin solution (5.0 μM) for 1 h. After being washed with PBS, the silicon wafer was co-incubated with 200 μL of PBS containing EpCAM and CKAAKN for 1 h and, then, it was washed and connected to the inlet and outlet Tygon fluid tubing to the 1-mL syringe (already installed on the syringe pump) for CTC capture.
Fabrication of microfluidic chip
In this work, the microfluidic system used a microfluidic piping (polydimethylsiloxane [PDMS]) system. By adopting the subtype structure, one entrance was gradually divided into eight parallel microfluidic channels (length 20 mm, wide 1 mm). There was a fishbone structure (with a width of 10 μm and a pitch of 10 μm) on the top of the microfluidic tubing. Height of the fishbone structure was 30 μm, and the overall height of the microfluidic tubing was 100 μm. PDMS chaotic mixers were fabricated based on a soft lithographic approach: 1) Designing and printing the masks of microfluidic channel structure and fishbone structure, respectively. 2) The microfluidic channel structure was fabricated by a standard photolithographic procedure; A negative photoresist (SU8-2100; MicroChem Corp., Newton, MA, USA) was spin-coated with a 70 μm thickness onto a 3 in silicon wafer. After exposure to UV under the masks of microfluidic channel structure, the microfluidic channel structure was obtained. 3). The fishbone structure: another negative photoresist (100 μm, SU8-2025; MicroChem Corp.) was spin-coated on the same wafer. Prior to UV irradiation, the mask of fishbone structure was aligned to get an accurate alignment between the prior pattern and the pattern to be imprinted. After exposure to UV under the masks of microfluidic channel structure, the fishbone structure was obtained. 4) The mold was then exposed to trimethylchlorosilane vapor for 2–3 min and then transferred to a Petri dish. 5) To prepare a 4.5 mm thick chip, a well-mixed PDMS prepolymer (RTV 615 A and B in 10–1 ratio; GE Silicones, Waterford, NY, USA) was poured into the mold and kept in an oven at 80°C for 48 h. The PDMS chaotic was then peeled off from the mold, and two through-holes were punched at the fabric channel’s ends for connection with the fluidic handler.
Preparation of artificial CTC sample
Cell suspensions (accurate counts) with a density of 1×104 cells/mL were prepared from BxPC3 culture dishes of non-small-cell pancreatic cancer cell lines with a culture abundance of about 95%. Then, through gradient dilution by PBS, BxPC3 cell suspensions with a density of 200/20 μL could be obtained. Afterward, 20 μL of BxPC3 cell suspensions (200 cells) was added into 180 μL of WBC suspensions (containing 2×105 cells), which was set as 200 μL of artificial CTC sample.
Procedure for CTC capture of artificial CTC sample
After completing the set-up of the microfluidic device, 200 μL of artificial sample (prepared freshly within 2 h) was injected into the system at different flow rates. As the completion of sample processing in the microfluidic chip, 100 μL of paraformaldehyde solution (2 wt%) was injected into the microfluidic chip (flow rate: 1.0 mL/h) to fix the captured cells. After disassembling the chips, the silicon nanowire substrate slide was removed and slightly washed with PBS. Then, 200 μL of the mixing solution of fluorophore labeled pancreatic cancer-specific markers of anti-CK (cytokeratin, a protein marker for epithelial cells) and anti-CD45 was dropped on the silicon nanowire substrate slide. After 24 h at 4°C, it was washed by PBS. Then, secondary antibody staining was carried out for 30 min, while the corresponding CK-PE antibody is the Alex488 marker and the corresponding CD45 antibody is the Alex555 marker. After cleaning, surface residue should be removed and, then, the silicon nanowire substrate slide was encapsulated by 80 μL of Hoechest solution (stained nuclear reagents). Fluorescence imaging could be performed by fluorescent microscope. The CK+/CD45−/DAPI+ cells were defined as CTC candidates; thus, the corresponding capture efficiency could be calculated (compared with initially added 200 cancer cells).
Procedure for CTC of clinical pancreatic cancer patients
A total of 16 pancreatic cancer patients diagnosed by pathologist from the Department of Oncology, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, and three healthy donors were enrolled. Written informed consent was obtained from all patients and healthy blood donors. Our project was approved by the ethics committee of The Central Hospital of Wuhan and was conducted in accordance with the Declaration of Helsinki. Taking 4 mL of whole blood from pancreatic cancer patients as an example, in our work, the method of gradient density centrifugation was adopted to preliminarily purify the blood of cancer patients. First, 4 mL of PBS was added into 4 mL of whole blood solution and mixed well. Then, the diluted blood sample was slowly added to a centrifuge tube (15 mL), while 4 mL of gradient density centrifugation solution (1,077) had been added into the tube already. Centrifugation was performed under 300× g for 40 min. Afterward, serum was removed and about 2–4 mL of peripheral blood mononuclear cells (PBMCs) was collected. Then, the collected PBMCs were further centrifuged at 400× g for 5 min. Afterward, the supernatant was removed and the PBMCs were washed with 2 mL of PBS. After the supernatant was removed by centrifugation, a mixture solution (400 μL) of freshly prepared anti-CK (5 μM), vimentin (VIM) (5 μM), and anti-CD45 (5 μM) was added. PBMCs were incubated for 45 min. After cleaning, they were set at 400 μL of PBS. Then, the CTC capture procedure and staining method were performed following the procedure of artificial CTC sample. Here, the CK+/CD45−/DAPI+ cells and VIM+/CD45−/DAPI+ cells were defined as CTC candidates to calculate the corresponding capture efficiency.
Cell viability assay for released cells
With the advantages of the high releasing efficiency presented by this method, the cell viability could be conveniently detected by the Vi-CELLTM XR (Cell Viability Analyzer; Beckman Coulter, Indianapolis, IN, USA). Following the capture procedure described earlier, the substrate-modified cancer cells are transferred into a 4°C refrigerator for 30 min. Then, the substrate was gently rinsed with a total of 0.5 mL of PBS for three times and the cell suspension solutions were collected. The released cancer cells on each silicon nanowires were finally prepared to a 1.0 mL volume cell suspension, which was then carefully transferred to a sample container especially for the Vi-CELLTMXR. The cell viability was analyzed as a triplicate.
Results and discussion
Preparation of microfluidic chip
Nanoscale structures were spread all over cellular surface, and there are many reports that prove the interactions between cells and nanostructures. By utilizing these interactions, many scientists adopted nanostructure to achieve efficient CTCs’ capture.26–29 Silicon nanowire, which were prepared through chemical wet etching process on silicon wafer (Figure 2), was therefore adopted and prepared the microfluidic device.30 First, a template was prepared on the surface of the silicon wafer (under the assist of Ag nanoparticle deposition) and then etched by chemical methods for patterning silicon nanowires. Generally, for the chemical etching step on silicon wafer, HF was often used.31 However, HF was very damaging to the operator and environment and would be the disadvantage for actual application. To address this issue, NH4F rather than HF was developed for etching in this work. The substitution of HF by NH4F was meaningful for operators that entire operation and preparation process could be completed in the general chemical laboratory. Meanwhile, the preparation process could be green for environment, thus enhancing the practicability. The obtained etched silicon wafer was characterized by scanning electron microscope (SEM). As shown in Figure 1B, it is clear that the surface of silicon wafer demonstrated typical nanowires. On the top of the device, there was staggered herringbone structures, which would enhance turbulence perpendicular to the silicon nanowires’ substrate, and thereby increase the interactions between cells and nanowires for further enhancement of CTCs capture efficiency.20,21 Meanwhile, microfluidic channels can also increase the sample transfer efficiency and speed up the sample processing, leading to high throughput. As a consequence, with environmentally friendly nanowires fabrication and new staggered herringbone structure, a new microfluidic chip was established to enhance CTCs capture efficiency.
Capture of CTCs with microfluidic device
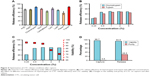
With the microfluidic chip in hand, it was then processed by chemical modification and biological functionalization to bond antibody for further CTC capture (detailed methods and procedures are seen in the Supplementary material). After the microfluidic device being ready, one typical pancreatic cancer cell of BxPC3 was chosen as an example to prepare artificial CTC sample (detailed procedures are described in the Supplementary material) to optimize test conditions and explore the performance of new microfluidic device. As described earlier, the immunoaffinity method of antibody was extensively adopted to capture CTCs, but single antibody usually lost intact CTCs. The strategy of combinatorial EpCAM and peptide of pancreatic cancer cell were therefore explored here. Through integrating EpCAM and peptide of CTCs would decrease the amounts of intact CTC and thus improve CTC capture efficiency. To verify our idea, the difference between the CTC capture efficiency of a single antibody and combination antibody with peptide at the same concentration was tested and calculated by counting the proportion of the cells captured on the microfluidic device (by the method of fluorescence staining, supporting information). As shown in Figure 3A, the capture efficiency by single EpCAM and CKAAKN were 77.8 and 74.8%, respectively, but after integrating them together (CKAAKN and EpCAM, abbreviate as CE cocktail), the capture efficiency of CE cocktail was greatly enhanced up to 95.6%, indicating the more precise capture of combinatorial EpCAM and CKAAKN.
Subsequently, suitable throughput of this microfluidic device was also investigated, following the procedures described in supporting information. As shown in Figure 3B, the microfluidic device realized the highest capture efficiency of 91.4%, when the flow rate is 1.0 mL/h. It is worth to note that even flow rate reached 5.0 mL/h and microfluidic device kept 82.6% capture efficiency, demonstrating its merit of high throughput. When the new microfluidic device realized excellent CTC capture, the possibility of false-positive results should be excluded. With this consideration, WBCs (2.0×106/mL) from healthy human blood, as reference, were adopted to prepare artificial blood sample without CTC. At the same time, the capture efficiency of different types of pancreatic cancer cells (BxPC3, Panc-1, and AsPC-1) were also explored. As shown in Figure 3C, with artificial blood sample without CTC, the capture efficient efficiency of WBCs was very low, indicating excellent anti-interference of this device. On the other hand, for different pancreatic cancer cells of BxPC3, Panc-1, and AsPC-1, our microfluidic device showed good stability that all of the three groups achieved high CTC capture efficiency. From these results, the positive and excellent CTC capture of this microfluidic device could be confirmed.
To clarify the contributions of different functional components in this device, the capture efficiencies of similar microfluidic devices with different functional components (Flat: device with flat surface; Flat/“CE cocktail”: device with flat surface and combinatorial EpCAM and CKAAKN; NWs: device with nanowires; and NWs/“CE cocktail”: device with nanowires’ surface and combinatorial EpCAM and CKAAKN) were compared and the results are shown in Figure 3D. For different types of pancreatic cancer cells, the microfluidic device with NWs/“CE cocktail” realized highest capture efficiency and then was NWs and Flat/“CE cocktail”, while the microfluidic device only with Flat demonstrated lowest capture efficiency. As compared to Flat device, higher capture efficiency of Flat/“CE cocktail” device could illustrated the enhanced CTC capture efficiency from combinatorial EpCAM and CKAAKN (the different results between NWs/“CE cocktail” device and NWs’ device could also prove this). On the other hand, NWs’ device showed higher capture efficiency than Flat device, confirming the advantages of nanowire structure. These results convinced us that both the designs in this microfluidic device of combinatorial EpCAM and CKAAKN and nanowires’ structure contributed to improve CTC capture efficiency. It is worth to note that the gap between NWs’ device and Flat device was much lower than that between NWs/“CE cocktail” device and Flat/“CE cocktail” device, providing the advantages of integration methods in microfluidic devices.
Release of CTCs in microfluidic device
To analyze CTCs, the captured CTCs should be released from microfluidic device. The purity and viability of obtained CTCs were very important for later analysis.8,14,15,32,33 Herein, enzymatic release of CTCs was investigated for the first time to achieve highly efficient release and minimal effect on cell viability. Because CTCs were captured by EpCAM, adding peptide digestion reagent to cut off the peptide should release CTCs from microfluidic device. In this work, nine different peptide digestion reagents (including ArgC, AspN, chymotrypsin, GluC, LysargiNase, LysC, LysN, pepsin, and trypsin) were adopted to explore their performance of CTC release in microfluidic device. As shown in Figure 4A, compared with other digestion reagents, chymotrypsin and trypsin illustrated much higher release efficiencies (85.5 and 86.2%, respectively). Subsequently, the CTCs’ release efficiencies of these two enzymes were further optimized by changing the concentration. According to Figure 4B, the best release efficiency (92.6%) of CTCs was realized at the concentration of 1 wt% by using chymotrypsin as the digestion reagent.
Cell viability of obtained CTCs was another key point for later analysis. High cell viability allowed in situ cell culture and analysis, which could simplify detection procedures. In this work, cell viability of released CTC with different concentrations of chymotrypsin was collected (the methods are described in the Supplementary material) and results are listed in Figure 4C. It is clear that cell viability of released CTCs was reduced, accompanying with the enhanced concentration of chymotrypsin. Considering the release efficiency and cell viability, 1 wt% chymotrypsin was the best condition for CTC release. Under the optimized release condition, the CTCs’ viability and purity (the “purity” is the percentage of CTCs accounts for the CTCs and immune cells; here, mainly refers to WBCs in peripheral blood) of captured and released were tested (Figure 4D). The results illustrated that this new microfluidic device realized high CTCs viability up to 96.7% but with low purity only 0.5% (due to nonspecific capture of WBCs by nanowire structure). After enzymatic release of chymotrypsin, CTCs still kept high viability 93.5%. Due to specific peptide digestion by chymotrypsin, CTCs were released from microfluidic device and achieved the good purity of 28.5%, which was high enough for later molecular biology analysis.
Detection of CTC of clinical pancreatic cancer patients
KRAS mutations are the hallmark of the late-stage pancreatic cancer, and KRAS also be considered as a key oncogene and therapeutic target in pancreatic cancer.34 After optimizing all the test conditions, this microfluidic device was further used for the detection of CTCs of actual clinical pancreatic cancer patients. Before the test, clinical blood samples should be pretreated firstly (the methods are described in the Supplementary material). During this process, clinical blood samples should be placed into BD Vacutainer Glass ACD Solution A tube (8.5 mL) to avoid the antigenic damage on the cell surface by EDTA anticoagulant (antigenic damage will affect the binding of antibody and reduce the capture efficiency). In addition, first 2 mL of blood sample in the tube should be removed, because there were exfoliated epithelial cells by needle, which might cause false-positive results.
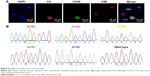
CTCs often have different cell phenotypes. Herein, the fluorescence imaging technology (the processing procedure is summarized in the Supplementary material) was employed to further verify and analyze different CTCs. As shown in Figure 5A, CTCs with different cell phenotypes were verified through unique fluorescence staining, in which DAPI was used to stain living cells, anti-CK was used to stain CTCs with epithelial cell phenotype, VIM was used to stain CTCs with interstitial cell phenotype,35 and CD45 was used to stain WBCs. This result confirmed our design that CTCs with different phenotypic ethnic groups could be captured by combinatorial EpCAM and CKAAKN for more effective CTC capture, which was significant in early warning of metastasis of pancreatic cancer.
After enzymatic release, harvested CTC still kept high viability and good purity, which could be used for the further test of CTC gene mutations in the blood. CTCs’ analysis from all pancreatic cancer patients and three healthy donors were carried out. Using the techniques described earlier, the released CTC suspensions were collected. After neutralization and washing, CTC pellets were subjected to DNA extraction and amplification using WGA4 single-cell whole genome amplification kit (Sigma-Aldrich Co., St Louis, MO, USA). Afterward, the pancreatic cancer marker gene KRAS was amplified and Sanger sequenced. As shown in Figure 5B, five types of KRAS mutation were detected in samples from 16 pancreatic cancer patients but not in healthy donors’ samples.
Conclusion
In this work, we have developed a simple assembly, easy to operate, and environmentally friendly microfluidic device Pe-SiNWS. The Pe-SiNWS could facilitate to specific capture, biological release, and precise analysis of CTC. Our work provided a novel method of CTC detection and would be a good candidate for actual clinical application.
Acknowledgments
We would like to thank the National Natural Science Foundation of China (81472748 and 81301084) and the fourth Huanghe Talents Plan for supporting this work.
Haitao Yang and Wenbo Wu are currently affiliated with the Department of Chemical and Biomolecular Engineering, National University of Singapore, Singapore.
Disclosure
The authors report no conflicts of interest in this work.
References
Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. | ||
Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039–1049. | ||
Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341(6151):1186–1188. | ||
Sarioglu AF, Aceto N, Kojic N, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12(7):685–691. | ||
Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253(2):180–204. | ||
Qian W, Zhang Y, Chen W. Capturing cancer: emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015;11(32):3850–3872. | ||
Nora Dickson M, Tsinberg P, Tang Z, Bischoff FZ, Wilson T, Leonard EF. Efficient capture of circulating tumor cells with a novel immunocytochemical microfluidic device. Biomicrofluidics. 2011;5(3):34119–3411915. | ||
Wang S, Liu K, Liu J, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50(13):3084–3088. | ||
Lin HK, Zheng S, Williams AJ, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16(20):5011–5018. | ||
Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192(3):373–382. | ||
Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5(179):179ra47. | ||
Moon HS, Kwon K, Kim SI, et al. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip. 2011;11(6):1118–1125. | ||
Rohani A, Moore JH, Kashatus JA, Sesaki H, Kashatus DF, Swami NS. Label-free quantification of intracellular mitochondrial dynamics using dielectrophoresis. Anal Chem. 2017;89(11):5757–5764. | ||
Shim S, Stemke-Hale K, Tsimberidou AM, Noshari J, Anderson TE, Gascoyne PRC. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics. 2013;7(1):011807. | ||
Zhang N, Deng Y, Tai Q, et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24(20):2756–2760. | ||
Shen Q, Xu L, Zhao L, et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013;25(16):2368–2373. | ||
Hou S, Zhao H, Zhao L, et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater. 2013;25(11):1547–1551. | ||
Hou S, Zhao L, Shen Q, et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed Engl. 2013;52(12):3379–3383. | ||
Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406. | ||
Shen Q, Peng C, Zhan Y. Aptamer–polymer functionalized silicon nanosubstrates for enhanced recovered circulating tumor cell viability and in vitro chemosensitivity testing. Int J Nanomed. 2016;11:2133–2146. | ||
Wang S, Liu K, Liu J, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50(13):3084–3088. | ||
Sheng W, Ogunwobi OO, Chen T, et al. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip. 2014;14(1):89–98. | ||
Valetti S, Maione F, Mura S, et al. Peptide-functionalized nanoparticles for selective targeting of pancreatic tumor. J Control Release. 2014;192(C):29–39. | ||
Liu H, Li Y, Sun K, et al. Dual-responsive surfaces modified with phenylboronic acid-containing polymer brush to reversibly capture and release cancer cells. J Am Chem Soc. 2013;135(20):7603–7609. | ||
Jeon S, Moon J-M, Lee ES, Kim YH, Cho Y. An electroactive biotin-doped polypyrrole substrate that immobilizes and releases EpCAM-positive cancer cells. Angewandte Chemie. 2014;126(18):4685–4690. | ||
Dang JM, Leong KW. Myogenic induction of aligned mesenchymal stem cell sheets by culture on thermally responsive electrospun nanofibers. Adv Mater. 2007;19(19):2775–2779. | ||
Park SY, Park SY, Namgung S, et al. Carbon nanotube monolayer patterns for directed growth of mesenchymal stem cells. Adv Mater. 2007;19(18):2530–2534. | ||
Jan E, Kotov NA. Successful differentiation of mouse neural stem cells on layer-by-layer assembled single-walled carbon nanotube composite. Nano Letters. 2007;7(5):1123–1128. | ||
Yang MT, Sniadecki NJ, Chen CS. Geometric considerations of micro- to nanoscale elastomeric post arrays to study cellular traction forces. Adv Mater. 2007;19(20):3119–3123. | ||
Gonchar KA, Zubairova AA, Schleusener A, Osminkina LA, Sivakov V. Optical properties of silicon nanowires fabricated by environment-friendly chemistry. Nanoscale Res Lett. 2016;11(1):357. | ||
Huang Z, Geyer N, Werner P, de Boor J, Gösele U. Metal-assisted chemical etching of silicon: a review. Adv Mater. 2011;23(2):285–308. | ||
Reátegui E, Aceto N, Lim EJ, et al. Tunable nanostructured coating for the capture and selective release of viable circulating tumor cells. Advanced Materials. 2015;27(9):1593–1599. | ||
Park MH, Reátegui E, Li W, et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J Am Chem Soc. 2017;139(7):2741–2749. | ||
Liu S, Tian Z, Zhang L, et al. Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget. 2016;7(37):59877–59891. | ||
Collins MA, Magliano MPD. Kras as a key oncogene and therapeutic target in pancreatic cancer. Front Physiol. 2013;4:407. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.