Back to Journals » International Journal of Nanomedicine » Volume 13
Biogenic pentagonal silver nanoparticles for safer and more effective antibacterial therapeutics
Authors Khan S, Ahmad K , Ahmad A, Raish M , Jan BL, Khan A, Khan MS
Received 15 March 2018
Accepted for publication 18 October 2018
Published 21 November 2018 Volume 2018:13 Pages 7789—7799
DOI https://doi.org/10.2147/IJN.S168224
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Salman Khan,1 Khurshid Ahmad,1 Ajaz Ahmad,2 Mohammad Raish,3 Basit L Jan,2 Altaf Khan,4 Mohd Sajid Khan1
1Nanomedicine & Nanobiotechnology Lab, Department of Biosciences, Integral University, Dasauli, Lucknow, India; 2Department of Clinical Pharmacy, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia; 3Department Pharmaceutics, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia; 4Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
Background: Biological synthesis of nanomaterials possesses unprecedented potential in the production of nanomaterials due to their ability to produce nanomaterials with improved biocompatibility in addition to eco-friendly synthetic procedures.
Methods: This article reports the isolation of an air-borne fungus from the campus of Integral University, Lucknow, with an exceptional ability to withstand very high concentrations of silver salt. The fungus was found to produce pentagonal silver nanoparticles (AgPgNps) when silver ions were reduced from silver nitrate. Molecular analysis and biochemical characterization techniques based on 18-seconds rRNA identified the fungus to belong to the Aspergillus sp. with the NCBI accession no KF913249. Material characterization techniques including ultraviolet (UV)–visible spectroscopy, transmission electron microscopy, and zeta potential analysis were used to satisfactorily characterize the as-synthesized AgPgNps.
Results: The AgPgNps synthesized by the fungus Aspergillus sp. exhibit an absorption that is maximum centered at about 416 nm, with a standard particle size of 23.22±2 nm. These AgPgNps exhibited broad-spectrum antimicrobial activities against an array of bacterial pathogens with remarkable minimum inhibitory concentration (MIC50) values: Staphylococcus aureus (ATCC 25923) – 9.230 µg/mL, Bacillus sp. (ATCC 14593) – 12.781 µg/mL, Escherichia coli (ATCC 25922) – 5.063 µg/mL, and Klebsiella pneumoniae (ATCC 13883) – 5.426 µg/mL. In vitro cytotoxicity analysis of biosynthesized AgPgNps showed a dose–response activity against human cervical cancer cell line (HeLa) and adenocarcinoma cells (A549) with MIC50 values of 0.038 µg/mL and 0.044 µg/mL, respectively.
Conclusion: These findings are very crucial to evaluate the biosynthetic process for the synthesis of nanoparticles (NPs) with unique properties. These NPs may find potential applications in sensing, medicine, and antimicrobial and anticancer therapies.
Keywords: pentagonal Ag NPs, fungus, biosynthesis, antibacterial agent, anticancer, in vitro cytotoxicity
Introduction
Different forms of silver such as silver metal, nitrates salts of silver, and silver sulfadiazine have been used to treat injuries due to burns and bacterial infections since ancient times.1 Silver metal also shows antimicrobial activity against a wide range of both Gram-negative and Gram-positive bacteria, fungi, protozoa, and certain viruses. The continuing fight against the emergence and robust nature of antibiotic-resistant bacteria has built high hopes over the efficacy of silver and silver-based compounds in tackling pathogens, including silver nanoparticles (NPs).2 Metallic silver in the form of silver NPs has shown lots of promise as a potential antimicrobial agent. Thus, a wide assortment of uses has discovered use of silver NPs going from silver-based dressings and silver-covered medicinal gadgets, eg, nanogels and nano lotions.1 The characteristics of microbicidal actions of silver NPs (from fungi to viruses) is an active field of research.3 Silver-based compounds such as silver-doped polymer fabrics, catheters, polyurethane,4 colloidal silver,5 fabrics laced with silver NPs,6 nano silver metal oxide granules,7 and ceramic materials coated with silver NPs8 are well known for their bactericidal functionality, while burn care uses silver compounds in an array of different forms.9 The bactericidal and veridical properties of silver and its derivatives have been altogether settled.10–12 However, in small concentrations, silver shows no toxicity toward animal cells, whereas it shows high toxicity toward microbes.
Silver has been widely applied in the medical industry since ancient times due to its inherent nontoxic nature.13,14 Silver possesses unique properties in that it is less toxic at certain concentrations to animal cells and certain bacteria while being effective against certain other pathogenic strains of bacteria at that concentration.15 It is widely established that the bactericidal properties of silver are ascribed to the catalytic oxidation by metallic silver and response with broke up univalent silver ions.16 The development of resistance by microbes against silver is not feasible since the microorganisms would have to undergo an array of mutations to overcome the damages to the targets attacked by the metals.13 In small concentrations, silver is unique in that it is safe for human cells at less concentrations while being very effective against different types of microbes and also has found application in medicines and in the maintenance of water and food qualities.17 In general, the toxic nature of metal NPs toward cells is attributed to their ability to produce reactive oxygen species (ROS).18 The ability of silver NPs to get oxidized slowly to release Ag+ ions renders them a potential biocide. In addition, these small-sized particles are ideal to move across the cell membrane to attack intracellularly. Additionally, the antibacterial properties of these NPs can also be attributed to their enhanced surface area to volume ratio characteristic.19 However, the effects of NPs on the inflammatory responses in humans have not been extensively investigated. Elechiguerra et al3 reported size-dependent interaction of silver NPs with HIV type 1 by binding to gp120 glycoprotein knobs to bring about an effect. In a similar note, they reported the size-dependent action of silver NPs against Gram-negative bacteria Escherichia coli.
In this study, a novel fungus was isolated from the campus of Integral University, Lucknow, and used to synthesize pentagonal silver nanoparticles (AgPgNps), which showed bactericidal properties against Bacillus spp. (ATCC 14593), Staphylococcus aureus (ATCC 25923), E. coli (ATCC 25922), and Klebsiella pneumoniae (ATCC 13883). These NPs showed no toxicity against primary osteoblast cells up to a moderately high concentration.
It might be estimated that different shaped (having almost equal surface area) silver NPs might be having distinctive features interacting with the surroundings. In this report, the relationship between the surface area of the NPs and their antimicrobial properties is not discussed. Even though there is a dearth of literature at present on the bactericidal efficacy of silver NPs related to their active surface areas, this study might serve as an important foundation to emphasize the shape-dependent bactericidal properties of silver NPs. Nonetheless, it is basic that the innovation in the methods of NP synthesis and their effect on different microbial strains should be investigated.13 Another study reported on the use of exopolysaccharides produced by Aeromonas punctata as the stabilizing agent by capping the NPs.21 A relatively less toxicity was shown by exopolysaccharide-capped particles toward S. aureus, E. coli, and Micrococcus luteus compared to that by non-capped particles, which shows that the encapsulation of exopolysaccharides acts as a physiological defense tool.22 Khan et al have done considerable research for the utilization of microbial-inferred biomolecules in the surface functionalization of nanomaterials. As indicated by their examination, zeta capability of NPs influences the surface adsorption of exopolysaccharides on NPs.2
Materials and methods
Materials
All the chemicals and solvents were bought from HiMedia, Merck, and Sigma-Aldrich Co. (St Louis, MO, USA).
Isolation of fungi
The fungus used in this study was isolated from the air in the campus of Integral University, Lucknow, Uttar Pradesh, India. Malt extract Glucose Yeast extract Peptone (MGYP) media containing 5 mM silver nitrate solution was exposed to the open air to entrap fungus capable of withstanding high concentrations of metal salt. The entrapped fungal strains were isolated to purity using the serial dilution method. The pure fungal strain was stored at −20°C in 30% glycerol.
Taxonomic identifications
The DNA was extracted from 48 hours old culture of fungus grown in 100 mL MGYP broth for phylogenetic analysis by analyzing 18S rRNA gene. Universal oligonucleotide primers 18SF (5′-CGW CGR AAN CCT TGT NAC GAS TTT TAC TN-3′) and 18 SR (5′-AWG CTA CST GGT TGA TCC TSC CAG N-3′) from Amnion Biosciences Pvt. Ltd. (Hyderabad, India) were used for amplification. The given polymerase chain reaction (PCR) program was followed for amplification of 18S rRNA gene (Bio-Rad thermal cycler; Bio-Rad Laboratories Inc., Hercules, CA, USA): one cycle of preheating for 5 minutes at 94°C and 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 3 minutes. These cycles were followed by incubation at 72°C for 5 minutes. Eventually, the reaction mixture was cooled to 4°C. The amplified PCR product was checked using 1% agarose gel with 5 kb molecular weight markers. The amplified gene was sequenced by the Sanger Dideoxy Sequencing/Chemistry-BDT version 3.1 method according to the company’s protocols using ABI 3730xl genetic analyzer (Amnion Biosciences Pvt. Ltd.). NCBI “BLASTn” program was used for alignment and assembly of the sequences. The neighbor-joining method was used to establish evolutionary history from 1,000 replicates.23
Microorganism and growth
The Potato Dextrose Agar slants with composition 20% w/v potato, 2% w/v dextrose, and 2% w/v agar were used to maintain new isolate of Aspergillus sp. (accession no KF913249) at 25°C. The 7-day-old Potato Dextrose Agar slant was used to inoculate a fungal mat (1 cm in diameter) into 100 mL liquid MGYP medium (malt extract [0.3% w/v], glucose [1.0% w/v], yeast extract [0.3% w/v], and peptone [0.5% w/v]) with incubation at 26°C±1°C on a rotary shaker (200 rpm) for 96 hours. Eventually, centrifugation (4,500× g, 20 minutes at 10°C) was used to collect mycelia under aseptic conditions with distilled water and used accordingly.
Extracellular synthesis of AgPgNps
The extracellular synthesis of AgPgNps was achieved by incubating mycelia (20 g wet mass) with 1 mM AgNO3 in 100 mL sterile distilled water under shaking conditions (200 rpm) for 120 hours at room temperature. The progress of synthesis of AgPgNps was checked regularly using ultraviolet (UV)–visible spectroscopy. The unutilized proteins were precipitated by using absolute ethanol (2 volume), and isolated AgPgNps were collected for further analysis.
Characterization of AgPgNps
Several physical techniques were used to characterize AgPgNps such as UV–visible spectroscopy, zeta potential analysis, dynamic light scattering, and transmission electron microscopy (TEM). Shimadzu dual-beam UV–visible spectrophotometer (model UV-1601 PC) operated on a declaration of 1 nm was used to authenticate the preliminary synthesis of AgPgNps. Zetasizer Nano-ZS model ZEN3600 (Malvern Instruments, Malvern, UK) was used to measure zeta potential. A drop of AgPgNps (suspension) was dried on a carbon-coated TEM copper grid to analyze the size of the inorganic core using transmission electron microscope (TEM) Tecnai™ G2 Spirit Bio-TWIN (FEI, Hillsboro, OR, USA) at a step-up voltage of 80 kV.
Fourier transform infrared (FT-IR) spectroscopy of biosynthesized AgPgNps
The presence of different functional groups at the surface of AgPgNps was confirmed by Perkin-Elmer Spectrum 2 FT-IR (PerkinElmer Inc., Waltham, MA, USA) furnished through a worldwide attenuated full reflectance sampling tool and scanned at room temperature using transmission approach above the wave number range of 4,000–650 cm−1 at a resolution of 4 cm−1.
Antibacterial activity
Determination of minimum inhibitory concentration (MIC50)
The preliminary antibacterial potential of AgPgNps was checked by the disk diffusion method.24 The MIC50 of confirmed antibacterial AgPgNps was evaluated by the microdilution method.25 Briefly, S. aureus (ATCC 25923), Bacillus sp. (ATCC 14593), K. pneumoniae (ATCC 13883), and E. coli (ATCC 25922) were obtained from NCIM, National Chemical Laboratory (Pune, India), developed to mid-logarithmic stage, gathered by centrifugation, rinsed with 10 mM sodium phosphate support (SPB) at pH 7.4, and diluted to 2×105 colony forming units (CFUs)/mL in SPB including 0.03% Luria-Bertani (LB) broth. The required concentrations (0.1–15 μg/mL) of AgPgNps with the bacterial inoculum (5×104 CFU per well) were successively diluted in 50 μL of LB medium within 96-well microtitre dishes and incubated overnight at 37°C. The lowest concentration of AgPgNps that inhibited 50% bacterial growth was considered as MIC50. The agar plate count method was adapted to further confirm the results. In this method, 25 μL aliquots from the freshly prepared 96-well microtitre plates (as abovementioned) were plated on LB plates and kept at 37°C for a night. Eventually, grown colonies of bacteria were counted on the next day.
Determination of minimum bactericidal concentration
S. aureus (ATCC 25923) – MIC50 9.230 μg/mL, Bacillus sp. (ATCC 14593) – MIC50 12.781 μg/mL, E. coli (ATCC 25922) – MIC50 5.063 μg/mL, and K. pneumoniae (ATCC 13883) – MIC50 5.426 μg/mL were used to evaluate minimum bacterial concentration (MBC). The culture (2 μL) from the well of a microtitre plate (which was used to determine MIC50) with no visible growth was transferred into another microtitre plate having 100 μL of broth for serial subcultivation and incubated for next 24 hours. The minimum concentration with no noticeable growth was characterized as MBC, showing a 99.5% loss of viability of the first inoculum. The bacterial growth was estimated at 620 nm wavelength using Microtitre Plate Manager 4.0 (Bio-Rad Laboratories Inc.) and evaluated with a blank. The same was also repeated on Petri plates for validity using dimethyl sulfoxide (DMSO) as the negative control. The experiment was repeated thrice for each compound with three replicates every time.
Cell culture
The human adenocarcinoma (A549) and human cervical growth cell line (HeLa) were received from National Center for Cell Science (NCCS; Pune, India). The primary osteoblasts were segregated from calvaria of the neonatal rat through enzymatic processing. Primary osteoblasts cells, A549 and HeLa, were developed as a monolayer in MEM, DMEM–F12, and EMEM media, individually, supplemented with 10% fetal bovine serum and 1% antibiotics containing 10,000 units of penicillin, 10 mg of streptomycin, and 25 μg of amphotericin B in a humidified CO2 incubator at 37°C. All the cell stocks were managed in 75 cm2 tissue culture flasks.
Assessment of cytotoxicity
The cytotoxicity of AgPgNps, A549, and HeLa cells was assayed according to the protocol described elsewhere26 where various concentrations (0.373, 0.186, 0.0932, 0.046, 0.0233, and 0.0116 μg/mL) of AgPgNps were used to determine the IC50 against each cell line.
Measurement of cytomorphological changes in A549 and HeLa cells
A549 and HeLa cells were pretreated with various concentrations of AgPgNps and kept warm for 48 hours at 37°C in 5% CO2 atmosphere. Toward the end of incubation, net morphological transforms in the cells were seen using an inverted phase contrast microscope (Nikon Eclipse Ti-S; Nikon Corporation, Tokyo, Japan).
Analysis of changes in nuclear morphology
Nuclear morphological changes in AgPgNp-treated and -untreated (control) primary osteoblast A549 and HeLa cells were observed under an Olympus fluorescence microscope associated with an Olympus Cool Snap Camera (Olympus Corporation, Tokyo, Japan) after staining with 4′,6-diamidino-2-phenylindole (DAPI). Observation of minimum 200 cells under the microscope revealed that live cells were having normal nuclei and blue chromatin with organized structure; stressed cells were found to have bright-blue chromatin, which was highly condensed, marginated, or fragmented. Three different softwares, namely, Micro-Image, ImageTool, and Matlab 7.0, were used for quantitative estimations. On the other hand, control DAPI staining revealed a single, bright, spherical nucleus and peripheral cell spots corresponding to stained mitochondria. The investigation of apoptosis on AgPgNp-treated A549 and HeLa cells were finished by staining with fluorescent nuclear dye DAPI. The cells were seeded and treated as mentioned earlier. Then, the cells were rinsed with PBS and kept in 4% paraformaldehyde for 10 minutes. Consequently, the cells were permeabilized through the permeabilizing buffer (3% paraformaldehyde and 0.5% Triton X-100) and stained with DAPI dye. A fluorescence microscope (Nikon Eclipse Ti-S) was used to acquire images of treated cells after staining. The cells with fragmented and condensed nuclei were measured as apoptotic cells.
Measurement of intracellular ROS generation
The Image-iT LIVE Green Reactive Oxygen Species Detection Kit (Invitrogen) was used to detect the production of ROS in AgPgNp-treated HeLa and A549 cells. The ROS production in viable cells was directly estimated by using a fluorogenic marker, 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA). The 96-well culture plates were used to seed the cells (1×104 per well) for 24 hours in a CO2 incubator at 37°C. Diverse concentrations (0.373, 0.186, 0.0932, 0.046, 0.0233, and 0.0116 μg/mL) of AgPgNps were used to incubate HeLa and A549 cells for 24 hours. Eventually, H2DCFDA (10 mM) was used to incubate cells for 30 minutes at 37°C. The reaction mixture was aspirated and changed by 200 μL of PBS in all the wells. The plates were kept back on a shaker for 10 minutes at room temperature within the dark. The intracellular fluorescence of cells was examined under an inverted fluorescence microscope (Nikon Eclipse Ti-S).
Quantitative estimation of ROS generation was done by re-seeding cells (1.5×104 per well) in 96-well black bottom culture plate with incubation for 24 hours at 37°C. Subsequently, cells were treated with various concentrations of AgPgNps (0.373, 0.186, 0.0932, 0.046, 0.0233, and 0.0116 μg/mL) for 12 hours and additionally incubated with 10 μM of DCFH-DA for 30 minutes at 37°C. Fluorescence intensity was estimated by the multiwall microplate reader (Synergy H1 Hybrid Multi-Mode Microplate Reader; BioTek, Winooski, VT, USA) at an excitation wavelength of 485 nm and the emission wavelength of 528 nm. Values were articulated as power percent overlay of fluorescence intensity with respect to control.
Results and discussion
Isolation, molecular characterization, and phylogenetic analysis of Aspergillus sp.
The fungus was isolated from the air at the campus of Integral University, Lucknow, India. In the quest for an uncommon fungus with uncommon features using an uncommon approach, MGYP media with 5 mM silver salt solution was exposed to the open air. In this particular media, only those fungi were able to grow that could withstand such high concentrations of silver ions. Furthermore, these fungi were found to possess the ability to produce AgPgNps by the reduction of silver ions.
The PCR-amplified 18S rRNA gene was found to be 1,718 bp long (Figure 1). This sequence was submitted to the NCBI with the unique accession number KF913249. The evolutionary history of the above gene was constructed based on the Jukes–Cantor model using the maximum likelihood method using Molecular Evolutionary Genetics Analysis 6 (Figure 2).27–29 A polyphasic approach combining phenotype and molecular techniques was used to classify taxonomy of the isolate. The sequence similarity of the 18S rRNA gene (1,718 bp) showed that this fungal isolate is a close relative of Aspergillus sp. This isolate encompasses a unique phylogenetic position with respect to Aspergillus fumigates and other species of Aspergillus sp. Thus, on the basis of physico-biochemical tests and molecular characteristics, this isolate appears to occupy a distinct phylogenetic position by being one of the unique species of Aspergillus and was named Aspergillus KF913249.
  | Figure 1 The Basic Local Alignment Search Tool analysis of the above sequence shows maximum similarity with Aspergillus terreus. |
Synthesis of pentagonal NPs
The AgPgNps were synthesized by incubating 100 g of fungal mycelia (in the log phase), Aspergillus sp. KF913249, in a solution of 1 mM AgNO3 in 100 mL distilled water at 27°C. It is proposed that Aspergillus sp. produces AgPgNps due to its ability to produce reducing enzymes with secondary metabolites, which could synergistically generate enough reducing potential to reduce AgNO3 (+1 oxidation state) into Ag (0 oxidation state) NPs in an aqueous medium. Reaction in the absence of salt confirmed the efficacy of mycelia in the synthesis of AgPgNps. Likewise, incubation of fungal mycelia alone in distilled water additionally did not create any assimilation peak(s) distinctive for AgPgNps. The characteristic absorption maximum (Figure 3A) centered at 416 nm attributed to the surface plasmon resonance (SPR) band of the AgPgNps. TEM analysis of the samples determined the average size of NPs to be 23±2 nm (Figure 3B). Further analysis of the TEM micrograph using Gatan DigitalMicrograph showed the pentagonal shape of NPs. The TEM micrographs showed distinct well-separated particles of as-synthesized AgPgNps. Zeta potential of AgPgNps was found to be −12.6±0.2 mV (Figure 3C), which made them stable for longer periods of time; the absence of aggregates is attributed to the electrostatic repulsive forces due to like forces presented by individual NPs.
Fourier transform infrared spectral analysis (Figure 3D) of AgPgNps presented peaks centered at 1,643.24 cm−1 characteristics of C=O of amide groups of the amide I linkage and amide II band presents as a medium-wide shoulder at 1,537.02 cm−1. The bands of amides I and II appear due to carboxyl stretch and N−H deformation vibrations in the amide bond of the proteins, which encapsulated AgPgNps.30 In addition, peak at 3,296.5 cm−1 confirms the N−H stretching vibration. This mode of vibration does not depend on the backbone conformation but is very sensitive to the strength of a hydrogen bond. Furthermore, a peak at 1,081.8 cm−1 shows (C−O−C/C−OH) C−O stretching30 of alcohol and ether group with C-N (aliphatic amine) stretching vibration. Due to the presence of various secondary metabolites, a peak at 2,127.39 cm−1 represents C=C stretch of alkynes. Terminal-free hydroxyl (O−H) is represented by representative peaks 3,756.013 cm−1 and 3,868.76 cm−1.
Antibacterial activity
In this study, the antimicrobial activity of AgPgNps was evaluated. The AgPgNps displayed microbicidal activity toward a range of different pathogenic microorganisms (Figure 4). The MIC50 values were determined against pathogenic bacteria, which included Bacillus sp. (12.781 μg/mL), S. aureus (9.230 μg/mL), E. coli (5.063 μg/mL), and K. pneumoniae (5.426 μg/mL), suggesting its broad-spectrum nature. This analysis further proved that AgPgNps displayed better efficacy toward Gram-positive strains as compared to the Gram-negative counterparts.
This analysis was performed by a method adapted from the method reported by Deda et al.31
Since ancient times, silver has enjoyed being the inorganic antibacterial agent of choice to fight against infections and spoilage. The antibacterial properties of silver NPs, which make it an ideal biocidal agent, are thought to be due to the slow oxidation exhibited by silver following the liberation of monovalent silver ion (Ag+) into the environment.19 Studies have well established the size-mediated antibacterial properties of silver NPs19,20 in detail, but very little is known about shape-dependent antimicrobial activity. However, shape-dependent mode of interaction of these NPs with the Gram-negative organism E. coli has been found to be dependent on the shape of NPs.13 In addition, some researchers have attempted to elucidate the mechanism responsible for the greater bactericidal property of NPs toward G− than that toward G+ bacteria. In general, a widely studied and accepted hypothesis is that generation of ROS is responsible for cellular toxicity exhibited by metal NPs (ROS).18 The microbicidal property exhibited by silver NPs has been widely believed to be due to loss of subcellular materials caused by the creation of pits in the cell membrane. This continues with the dehydrogenases involved in the respiratory chain getting inhibited, affecting the growth of cells altogether. Meanwhile, some proteins and phospholipids may work together to initiate the collapse of the membrane, eventually leading to cell decomposition and death.32 The Gram-positive bacterial cells respond differently following exposure to metal NPs.32–35 The most recent mechanism states that silver NPs target both the secondary (α-helix) and primary structures of the bacterial cell wall, by making connections on both the surface of the peptide and glycan ports of a bacterial cell wall, leading to the generation of “pits” on it. Silver NPs connect to β–1/4 bonds of N-acetylglucosamine and N-acetylmuramic acid of glycan strands, destroying their linkage and releasing them into the growing environment.36
The cytotoxic effect of AgPgNps was tested on A549 and HeLa cell lines using MTT assay where primary osteoblasts were taken as control. The viability of these cells was measured at various concentrations (0.373, 0.186, 0.0932, 0.046, 0.0233, and 0.0116 μg/mL) of AgPgNps (Figure 5). The results clearly illustrated that HeLa cells (Figure 5) rapidly lost their viability in comparison to A549 cells when incubated with AgPgNps in the concentration range of 0.0116–0.046 μg/mL, which was trailed by an attenuated decrease when the NPs were used at higher concentrations from 0.046 to 0.373 μg/mL. Interestingly, these NPs were found to be less cytotoxic to primary osteoblasts (Figure 5), resisting to a higher permissible biological limit.
In addition, when 70% confluent HeLa and A549 cells were incubated with AgPgNps for 48 hours, the phase-contrast microscopic images (Figure 6) showed changes in cellular morphology where majority of the cells of HeLa (Figure 6B) and A549 (Figure 6D) experienced changes in shape turning circular, with failure of membrane integrity, inhibition of cell growth, condensation of cytoplasm, and clumping of cells. On the other hand, unimportant changes in morphology were observed for primary osteoblasts (picture not shown) and untreated cells (Figure 6A and C). The quantification of these morphological changes was performed by means of ImageTool software. The extent of roundedness of the cell was quantified to measure the change in the morphology of the cells. Furthermore, the mechanism of cellular uptake and internalization of AgPgNps was analyzed using a fluorescent stain DAPI. Caveolae-mediated endocytosis is implicated in the internalization of anionic AgPgNps. These are microdomains produced in the caveolar vesicles that are generally made up of sphingomyelins (an amine group present on its polar domain), cationic lipids that intermingle and initiate the endocytosis of anionic NPs. Furthermore, NPs might also enter cells non-specifically through macropinocytosis, which is a vesicular transmonolayer transport that involves dynamin and microtubule network. AgPgNPs internalized through caveolae-mediated endocytosis enjoy improved bioavailability since they are prevented from getting degraded by avoiding interaction with lysosomes after their entry. This improved the efficacy of these NPs by enabling them to interact with subcellular structures further down. After entering into cells, AgPgNps containing vesicles fused with caveosomes or multivesicular body. Subsequently, caveosomes containing AgPgNps moved along with microtubules to the endoplasmic reticulum (ER),35 eventually leading to their entry into the nucleus through nuclear pore complex.37,38 In addition, clathrin–caveolae-independent endocytosis involved Arf6-, Cdc42-, and Rhoa-dependent pathways39 and cholesterol with specific lipid compositions. Hence, AgPgNps causes apoptosis by condensing chromatin, which can be observed by DAPI analysis.
It is a well studied and accepted fact that pro-inflammatory and toxic activities of inorganic NPs including silver NPs are brought through the production of ROS. When human cells are exposed to oxidative stress, they result in an increase in the transcription of proinflammatory cytokines inducing apoptosis brought about by the c-Jun N-terminal kinase pathway.40,41 The silver NPs are known to be cytotoxic, genotoxic, and antiproliferative because ideally, DNA-damaging agents induce genome instability, which is a predisposing factor in carcinogenesis.
AgPgNps augmented intracellular ROS generation in HeLa and A549 cells
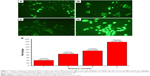
The amount of intracellular ROS generated actually dictates the caspase-dependent and caspase-independent cell death. The production of intracellular ROS has been implicated in cell death induced by both caspase-dependent and independent forms.42,43 In order to corroborate the mechanism of action of AgPgNps, the intracellular ROS levels of both AgPgNp-treated and -untreated HeLa (Figure 7A and B) and A549 cells (Figure 7C and D) were determined by the DCFH-DA method. Briefly, HeLa and A549 cells were monitored for changes in intracellular ROS levels by staining them with DCFH-DA after12 h of treatment. AgPgNp-treated cells (Figure 7A) demonstrated more grounded DCF-fluorescence intensity in both HeLa and A549 cells, which suggested an upgraded intracellular ROS level (Figure 7A). The above perceptions were further approved by the quantitative estimation of the ROS level, which demonstrated that intracellular ROS level was raised by approximately 2.76 folds (HeLa cells) and 2.01 folds (A549) when contrasted with control when both HeLa and A549 cells were treated with 0.046 μg/mL of AgPgNps separately. Consequently, both the above outcomes recommended that AgPgNps enhanced ROS development in both the cell lines in a dose-dependent manner (Figure 7E).
Conclusion
We report the isolation of a fungus from Integral University campus and characterization using molecular and biochemical methods. This strain of fungus is capable of withstanding high concentrationd of metal ions. Hence, this fungus was used for the production of AgPgNps. These biogenic NPs showed effective antibacterial activity and anticancer activity in a dose-dependent manner. Additional researches are expected to evaluate the toxicity in detail and the mechanism implicated in the antibacterial and anticancer potential of the synthesized AgPgNps, which may propel these NPs as a potent nanomedicine.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding through research group number (RG-1438-069). The authors acknowledge Integral University and Department of Biosciences for providing facilities and support for the research.
Disclosure
The authors report no conflicts of interest in this work.
References
Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27(1):76–83. | ||
Ravindran A, Chandran P, Khan SS. Biofunctionalized silver nanoparticles: advances and prospects. Colloids Surf B Biointerfaces. 2013;105:342–352. | ||
Elechiguerra JL, Burt JL, Morones JR, et al. Interaction of silver nanoparticles with HIV-1. J Nanobiotechnology. 2005;3(1):6–10. | ||
Cho JW, So JH. Polyurethane–silver fibers prepared by infiltration and reduction of silver nitrate. Mater Lett. 2006;60(21–22):2653–2656. | ||
Prucek R, Kvítek L, Hrbáč J. Facultas rerum naturalium. Chemica. 2004;43:59. | ||
Yan J, Cheng J; CC Technology Investment Co., Ltd., Hong Kong (CN). Antimicrobial Yarn having nanosilver particles and methods for manufacturing the same. United States patent US 6,979,491 B2. 2005 Dec 27. | ||
Yan J, Cheng J. GlobeAsia, L.L.C Hanover, MD (US). Nanosilver-Containing antibacterial and antifungal granules and methods for preparing and using the same. United States patent US 6, 379, 712 B1. 2002 Apr 30. | ||
Kawasumi S, Yamada M, Honma M. Kawasumi Laboritories, Inc,m Kanagawa, Japan. Metallic bactericidal Agent. United States patent 5, 824, 267. 1998 Oct 20. | ||
Klasen HJ. Historical review of the use of silver in the treatment of burns. I. Early uses. Burns. 2000;26(2):117–130. | ||
Oka H, Tomioka T, Tomita K, Nishino A, Ueda S. Inactivation of enveloped viruses by a silver-thiosulfate complex. Met Based Drugs. 1994;1(5–6):511. | ||
Oloffs A, Grosse-Siestrup C, Bisson S, Rinck M, Rudolph R, Gross U. Biocompatibility of silver-coated polyurethane catheters and silver-coated Dacron material. Biomaterials. 1994;15(10):753–758. | ||
Tokumaru T, Shimizu Y, Fox CL. Antiviral activities of silver sulfadiazine and ocular infection. Res Commun Chem Pathol Pharmacol. 1984;8:151–158. | ||
Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol. 2007;73(6):1712–1720. | ||
Klueh U, Wagner V, Kelly S, Johnson A, Bryers JD. Efficacy of silver-coated fabric to prevent bacterial colonization and subsequent device-based biofilm formation. J Biomed Mater Res. 2000;53(6):621–631. | ||
Zhao G, Stevens SE. Multiple parameters for the comprehensive evaluation of the susceptibility of Escherichia coli to the silver ion. Biometals. 1998;11(1):27–32. | ||
James GV. Water treatment. 4th ed. Cleveland, OH: CRC Press; 1971:38. | ||
Batarseh KI. Anomaly and correlation of killing in the therapeutic properties of silver (I) chelation with glutamic and tartaric acids. J Antimicrob Chemother. 2004;54(2):546–548. | ||
Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. | ||
Krutyakov YA, Kudrinskiy AA, Olenin AY, Lisichkin GV. Synthesis and properties of silver nanoparticles: advances and prospects. Russ Chem Rev. 2008;77(3):233–257. | ||
Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346–2353. | ||
Sudheer Khan S, Mukherjee A, Chandrasekaran N. Interaction of colloidal silver nanoparticles (SNPs) with exopolysaccharides (EPS) and its adsorption isotherms and kinetics. Colloids Surf A Physicochem Eng Asp. 2011;381(1–3):99–105. | ||
Sudheer Khan S, Bharath Kumar E, Mukherjee A, Chandrasekaran N. Bacterial tolerance to silver nanoparticles (SNPs): Aeromonas punctata isolated from sewage environment. J Basic Microbiol. 2011;51(2):183–190. | ||
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. | ||
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. | ||
Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in Laboratory Medicine. Baltimore: Williams & Wilkins; 1991:72–78. | ||
Iram S, Zahera M, Khan S, et al. Gold nanoconjugates reinforce the potency of conjugated cisplatin and doxorubicin. Colloids Surf B Biointerfaces. 2017;160:254–264. | ||
Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969:21–132. | ||
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. | ||
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. | ||
Huang J, Li Q, Sun D, et al. Biosynthesis of silver and gold nanoparticles by novel sundried Cinnamomum camphora leaf. Nanotechnology. 2007;18(10):105104–105111. | ||
Deda DK, Cardoso RM, Uchiyama MK, et al. A reliable protocol for colorimetric determination of iron oxide nanoparticle uptake by cells. Anal Bioanal Chem. 2017;409(28):6663–6675. | ||
Li WR, Xie XB, Shi QS, Zeng HY, Ou-Yang YS, Chen YB. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol. 2010;85(4):1115–1122. | ||
Sondi I, Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Colloid Interface Sci. 2004;275(1):177–182. | ||
Yamanaka M, Hara K, Kudo J. Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl Environ Microbiol. 2005;71(11):7589–7593. | ||
Vaidyanathan R, Gopalram S, Kalishwaralal K, Deepak V. Ram Kumar Pandian S, Gurunathan S. Enhanced silver nanoparticle synthesis by optimization of nitrate reductase activity. Colloid Surf B. 2010;75:335–341. | ||
Mirzajani F, Ghassempour A, Aliahmadi A, Esmaeili MA. Antibacterial effect of silver nanoparticles on Staphylococcus aureus. Res Microbiol. 2011;162(5):542–549. | ||
Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58(1):32–45. | ||
Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Annu Rev Microbiol. 1998;52:627–686. | ||
Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8(8):603–612. | ||
Rinna A, Magdolenova Z, Hudecova A, Kruszewski M, Refsnes M, Dusinska M. Effect of silver nanoparticles on mitogen-activated protein kinases activation: role of reactive oxygen species and implication in DNA damage. Mutagenesis. 2015;30(1):59–66. | ||
Ma J, Zhao D, Lu H, Huang W, Yu D. Apoptosis signal-regulating kinase 1 (ASK1) activation is involved in silver nanoparticles induced apoptosis of A549 lung cancer cell line. J Biomed Nanotechnol. 2017;13(3):349–354. | ||
Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. | ||
Wenzel U, Nickel A, Daniel H. alpha-Lipoic acid induces apoptosis in human colon cancer cells by increasing mitochondrial respiration with a concomitant O2-*-generation. Apoptosis. 2005;10(2):359–368. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.