Back to Journals » Clinical Interventions in Aging » Volume 10
Associations between apolipoprotein E gene polymorphisms and Alzheimer’s disease risk in a large Chinese Han population
Authors Wu P , Li H, Liu Z, Tao Q, Xu M, Guo Q, Hong Z, Sun Y
Received 28 August 2014
Accepted for publication 3 November 2014
Published 30 January 2015 Volume 2015:10 Pages 371—378
DOI https://doi.org/10.2147/CIA.S73396
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Zhi-Ying Wu
Ping Wu,1,2 Hong-Lei Li,1 Zhi-Jun Liu,1 Qing-Qing Tao,1 Miao Xu,1 Qi-Hao Guo,1 Zhen Hong,1 Yi-Min Sun1
1Department of Neurology and Institute of Neurology, 2PET Center, Department of Nuclear Medicine, Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China
Objective: Apolipoprotein E gene (APOE) polymorphisms contributing to the risk of sporadic Alzheimer’s disease (AD) have been identified for decades, but it has not been investigated in large AD samples of Chinese Han population.
Methods: We performed a cross-sectional study to explore the effect of APOE polymorphisms on sporadic AD in 875 sporadic AD patients and 1,195 cognitive normal controls of Chinese Han. Genotyping of APOE was determined by multiplex amplification refractory mutation system polymerase chain reaction.
Results: APOE ε3ε4 and ε4ε4 genotypes increased AD risk with dosage effect. The odds ratio (OR) of ε3ε4 was 1.89 and the OR of ε4ε4 was 15.64 compared with that of ε3ε3 in all the subjects. E2ε3 genotype decreased AD risk in all the subjects (OR=0.64), female subgroup (OR=0.57), and late-onset AD subgroup (OR=0.60). However, neither ε2ε2 nor ε2ε4 affected AD risk. About the age at onset (AAO), the influence of APOE ε4 was only exhibited in late-onset AD subgroup, with 1 year lower in ε4-positive ones than negative ones. Further analysis did not show the dosage effect of Ε4 pertinent to AAO, though the AAO of ε4ε4 patients decreased by 2 years. E2 did not affect the AAO of AD.
Conclusion: APOE ε4 is a strong risk factor of AD risk in Chinese Han population, and APOE ε4ε4 genotype might be related to the AAO of late-onset AD.
Keywords: sporadic, cross sectional study, dosage effect, age at onset
Introduction
Alzheimer’s disease (AD) is the most common cause of senile dementia characterized by progressive decline in cognition and behaviors. The cause of AD was complex, and genetic factors contributed to its risk. Mutations on three genes of amyloid precursor protein, presenilin 1, and presenilin 2 are associated with rare familial early-onset AD (EOAD). As for the majority of sporadic AD (SAD), apolipoprotein E gene (APOE) was the only one confirmed to be related with SAD risks since 1993,1,2 and the results were replicated by many candidate genetic studies in different populations and different regions all around the world.3 Recent genome-wide association studies found that APOE is far more significantly related to AD risk than all the other candidate loci.4,5
The APOE gene located in 19q13.2, encoding apoE, which consists of 299 amino acids, is a cholesterol carrier involved in lipid transportation and injury repair in the brain. APOE has three common isoforms termed ε2, ε3, and ε4, which could be determined by cysteine–arginine substitutions at residues 112 and 158. The frequencies of these three alleles are different among ethnics.6 Generally, ε3 is the most common allele, accounting for 60%–90% of the allelic variation. The ε2 constitutes 0%–20% of allelic variation and ε4 constitutes 10%–20% (http://asia.ensembl.org/Homo_sapiens/Variation/Population). The ε4 was proved to increase AD risk in a dose-dependent pattern and lower the age at disease onset compared with ε4 noncarriers.7 It was also reported as a risk factor for the conversion of mild cognitive impairment to AD.8 In contrast, ε2 was reported to have a “protective” effect on AD risk and to slower cognitive function decline than ε2-negative status.9
The association between APOE polymorphisms and AD risk has been investigated in Caucasian, Hispanic, African American, Japanese, and small numbers of Chinese Han populations.10–12 Other studies of candidate genes and AD risk in Chinese Han population used APOE ε4–carrying status as a stratification sign but did not focus on APOE itself.13 Here we investigated a large number of 875 SAD and 1,195 controls of Chinese Han to explore the association between APOE polymorphisms and AD risk.
Materials and methods
Subjects
A total of 875 SAD patients and 1,195 unrelated healthy controls were included in this cross-sectional study. All the subjects were from Chinese Han population. SAD patients were recruited from memory disorders clinics in Huashan Hospital between March 2007 and September 2013 with a median age of 72 years (range 48–100 years). Cognitively normal controls with age, sex, and origins similar to SAD patients were recruited from the community epidemiologic investigations (median age of 69 years, range 48–94 years). The enrollment procedure and the inclusion and exclusion criteria for SAD cases and controls were as previously reported.14 The diagnosis of AD was according to the criteria of Diagnostic and Statistical Manual of Mental Disorders IV revised. Written consents were obtained from subjects or their legally authorized caregivers. This study was approved by the ethics committee of Huashan Hospital.
Genotyping of APOE
Genomic DNA was extracted from peripheral blood using a Blood Genomic DNA Extraction Kit (TIANGEN, Shanghai, People’s Republic of China). The APOE genotypes were determined by multiplex amplification refractory mutation system polymerase chain reaction according to the method previously described.15
Statistical analysis
Hardy–Weinberg equilibrium tests of APOE polymorphisms within the groups were performed using χ2 analysis. The χ2 test or Student’s t-test was used to test for the differences between AD and control subjects in the distribution of sex, age at onset (AAO), and mini-mental state examination scores. The χ2 test was used to compare the genotypes and allele frequencies between AD patients and control subjects. Odds ratio (OR) and the 95% confidence interval (CI) for testing possible associations between AD and control groups were determined by binary logistic regression analyses; AAO and sex were used as covariates. The potential effects of each genotype on AAO in AD patients were calculated by one-way analysis of variance and further analysis by post hoc least significant difference. All statistical analyses were performed using SPSS version 13.0 (SPSS Inc, Chicago, IL, USA). P<0.05 was considered significant.
Results
General information
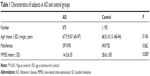
General information of the participants is shown in Table 1. No significant difference was found in age and sex between the two groups, while the mini-mental state examination score was significantly lower in AD. The distributions of the six common genotypes of APOE were under Hardy–Weinberg equilibrium in SAD patients and control subjects, respectively (Table S1).
ε2 allele decreased AD risk and ε4 allele increased AD risk
In all the subjects, the distribution of allele frequencies and genotypes of APOE was of significant difference between AD and control groups with more ε2, ε3 allele and less ε4 allele in controls (Table 2). There were also more ε2-carrying subjects (ε2ε2, ε2ε3, and ε2ε4) and less ε4-carrying subjects (ε2ε4, ε3ε4, ε4ε4) in the control group than in the AD group. When the subjects were further stratified by sex and AAO (AD with AAO ≤65 was defined as EOAD; AAO >65 as late-onset AD [LOAD]), the differences remained significant (Table 2).
The impact of APOE genotype and allele frequencies on SAD risks was analyzed by binary logistic regression. As shown in Table 3, in all the subjects, ε2ε3 genotype decreased AD risk (P=8×10-3, OR 0.64, 95% CI 0.46–0.89) while ε2ε2 and ε2ε4 genotypes did not statistically relate to AD risk. On the contrary, ε3ε4 and ε4ε4 genotype increased AD risk with dosage effect of ε4: the OR of ε3ε4 was 1.90 (P=1.18×10-9, 95% CI 1.54–2.33) while the OR of ε4ε4 rose to 15.64 (P=8.59×10-15, 95% CI 7.92–32.05). When the subjects were further divided by sex and AAO, the ε4 dosage effect remained constant, but the protective effect of ε2ε3 was significant only in the female subgroup and LOAD subgroup. As for the ε2- and ε4-carrying status, ε2 allele lowered the risk of developing AD while ε4 increased this risk, which existed in all subgroups after the subjects were stratified by sex and AAO, with the highest OR of 2.79 in female ε4-positive subjects (Table 3).
APOE ε4ε4 may be associated with an earlier AAO in LOAD patients
In AD patients, only in LOAD was the AAO found to be significantly lower in ε4 allele-positive subjects than ε4 allele-negative ones (73.9±5.12 vs 74.9±5.18, P=2.2×10-2) (Table 4). Nevertheless, there was no difference in AAO between the ε2-positive AD and ε2-negative ones. We further investigated the AAO according to the dosage of ε2 and ε4. The AAO was 2 years lower in patients of ε4ε4 genotype than ε4 carriers (ε2ε4 and ε3ε4) or ε4-negative ones. But no dosage effect of ε4 on AAO was found.
We further investigated the effect of APOE ε2 and ε4 haplotype on AD risk in different AAO and found ε2’s protective role against AD in the patients with AAO of 61–65 and above 76 (Table 5). The ε4 haplotype increased AD risk in patients with AAO below 55 and 61–75 with the highest OR of 3.842 in 66–70 groups. The risk decreased when AAO was above 76, though there was no significant difference.
Discussion
In 1993, APOE ε4 was first reported to increase SAD risks and advance AAO of AD in a gene dosage way.1,2,16 Except for several investigations,17 most studies confirmed the results.18–23 The dose effect of APOE ε4 on AD risk was reported to be ascribed to increased Aβ, Aβ oligomers, and plaque deposition and reduced metabolism in certain parts of the brain.24
The allele frequencies and genotypes vary among different ethnic groups.25 In our study, the APOE allele frequencies (AD: ε2 4.8%, ε3 68.1%, and ε4 27.1%; control ε2 8.5%, ε3 78.1%, and ε4 13.5%) were similar to the APOE survey in Shanghai consisting of 65 AD patients and 363 cognitively normal controls (AD: ε2 4.6%, ε3 70%, and ε4 25.4%; control: ε2 8.6%, ε3 80.4%, and ε4 11%).6 We also found that the APOE ε4 was the independent risk factor of AD and increased the AD risk in a gene dosage way. This is quite consistent with the investigations in other populations.7,26,27 However, the ORs in ε3ε4 genotype toward AD risks were relatively smaller in all the subjects and subgroups of LOAD, EOAD, male, and female (1.579–2.159) groups, compared with those reported in other studies7,21,25,28 (usually >2). The ORs in ε4ε4 genotype ranged from 11.061 to 20.581, much greater than those of ε3ε4 and in accordance with the results in other studies. The genotype of ε2ε4 showed a risk factor of AD in some studies,7 but did not show any protective or risk effect on AD pathogenesis in our study, which might be due to the small frequency of ε2ε4 genotypes or the co-existence of a “protective” and harmful effect in ε2 allele and ε4 allele, respectively.
In the present investigation, the APOE ε2 decreased the AD risks, and ε2ε3 lowered the AD risk in total population, female, and LOAD subgroups. This effect is similar to those observed in other populations.27,29 But the “protective” effect did not always show due to the lower frequency of ε2 allele.7,30 In one study of north Chinese population, the ε2 was indicated as a protective factor in male population, which was contrary to our result. This might be contributed to their small subject number and imbalanced sex distribution between AD and control groups.12
Researches indicated that the ε4 allele took part in the pathogenesis of EOAD as well,29,31 and it was well replicated in our study. But a previous report in Chinese population did not find any association, which could be due to the small number of subjects.12 Some investigations indicated that ε4 increased AD risk in women more than men,32,33 but others did not find the pattern.21,29 In a prospective study in Latin Americans, APOE ε4 allele risk was significant only in women,32 but had a stronger effect in men from Sweden and Finland.27,34 In our population, APOE ε4 increased AD risk in both sexes with a higher OR in the female group. The different results might be caused by ethnic origins.
APOE ε2 allele was reported not to affect AAO in AD patients in most studies.31 The differences of AAO between ε2ε2 group and one or no ε2 group did not reach significant, though the AAO of ε2ε2 was 4 years later in number than that of one ε2 group. This might be attributed to the very low frequencies of ε2ε2 genotypes in AD. APOE ε4 allele was reported to lower the AAO of LOAD in a gene dosage way. The AAO of AD patients with APOE ε4ε4 genotype was 5–16 years lower than those with ε4-negative ones.16 In our subjects, the AAO had a decrease of 2 years in patients of ε4ε4 genotype in contrast to ε4 carriers (ε2ε4 and ε3ε4) or ε4-negative ones, but not in a gene dosage way. Though we found some significant differences in AAO in the current study, we still could not deduce the effect of APOE genotype on the AAO with respect to the cross-sectional study. So further prospective studies should be implemented in Chinese Han population.
Whether APOE ε2 would reduce AD risk in a certain AAO range was controversial.27,35 We found ε2’s protective role in the AAO of 61–65 and above 76. As for ε4 haplotype, its AD risk decreased in people aged 70 years and above,36 which was quite similar to our results. There might be other genetic risk factors or environmental effects contributing to the AD onset in very old people.
This study had a few limitations. First, factors like diabetes, history of depression, stroke, and heart attack may also contribute to AD pathogenesis, but due to the incomplete information, we did not put those covariants into the binary logistic regression. Second, the subjects were recruited from memory disorders clinics but not the general population, which might exaggerate the impact of APOE ε4 on AD risks. In the population-based studies, the positive predictive value was lower, and hence APOE ε4 is not recommended for a screen test for AD.22,34
In conclusion, the APOE ε4 allele is a strong risk factor in AD in the Chinese Han population and it affects AD risk in a gene dosage effect, similar to those in other populations. The risk of APOE ε3ε4 toward AD was relatively smaller and the APOE ε4ε4 genotype lowered the AAO of AD in the LOAD group, which might be the specific characteristics of APOE polymorphisms in the Chinese Han population. The role of APOE protein in AD pathogenesis and other loci that will help to increase the AD-predictive value combined with APOE ε4 should be studied in the future.
Acknowledgments
The authors thank the participants of this study for their cooperation. This work was supported by a grant from the National Natural Science Foundation of China to Yi-Min Sun (81401048).
Author contributions
Ping Wu was responsible for data collection, data analysis, and manuscript drafting. Yi-Min Sun was responsible for study design, data analysis, data interpretation, and editing the manuscript. Hong-Lei Li, Zhi-Jun Liu, Qing-Qing Tao, Miao Xu, Qi-Hao Guo, and Zhen Hong were responsible for study implementation and data collection. All authors contributed equally to revising the manuscript and have approved the final version.
Disclosure
The authors report no conflicts of interest in this work.
References
Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. | ||
Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. | ||
Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. | ||
Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. | ||
Naj AC, Beecham GW, Martin ER, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6:e1001130. | ||
Katzman R, Zhang MY, Chen PJ, et al. Effects of apolipoprotein E on dementia and aging in the Shanghai Survey of Dementia. Neurology. 1997;49:779–785. | ||
Sando SB, Melquist S, Cannon A, et al. APOE epsilon 4 lowers age at onset and is a high risk factor for Alzheimer’s disease; a case control study from central Norway. BMC Neurol. 2008;8:9. | ||
Fei M, Jianhua W. Apolipoprotein epsilon4-allele as a significant risk factor for conversion from mild cognitive impairment to Alzheimer’s disease: a meta-analysis of prospective studies. J Mol Neurosci. 2013;50:257–263. | ||
Bonner-Jackson A, Okonkwo O, Tremont G. Apolipoprotein E epsilon2 and functional decline in amnestic mild cognitive impairment and Alzheimer disease. Am J Geriatr Psychiatry. 2012;20:584–593. | ||
Zhou X, Miao H, Rausch WD, et al. Association between apolipoprotein E gene polymorphism and Alzheimer’s disease in Uighur and Han populations. Psychogeriatrics. 2012;12:83–87. | ||
Lai S, Chen Y, Wen Z. Association between apolipoprotein E polymorphism and Alzheimer’s disease: a population-based study in Guangzhou, China. Zhonghua Liu Xing Bing Xue Za Zhi. 2001;22:202–204. | ||
Chen D, Zhang JW, Zhang ZX, et al. [Apolipoprotein E gene polymorphisms and Alzheimer disease]. Yi Chuan Xue Bao. 2003;30:1167–1170. | ||
Liu G, Zhang L, Feng R, et al. Lack of association between PICALM rs3851179 polymorphism and Alzheimer’s disease in Chinese population and APOE epsilon4-negative subgroup. Neurobiol Aging. 2013;34:1310–1319. | ||
Sun YM, Li HL, Guo QH, et al. The polymorphism of the ATP-binding cassette transporter 1 gene modulates Alzheimer disease risk in Chinese Han ethnic population. Am J Geriatr Psychiatry. 2012;20:603–611. | ||
Donohoe GG, Salomaki A, Lehtimaki T, Pulkki K, Kairisto V. Rapid identification of apolipoprotein E genotypes by multiplex amplification refractory mutation system PCR and capillary gel electrophoresis. Clin Chem. 1999;45:143–146. | ||
Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. | ||
Poduslo SE, Huang R, Huang J, Smith S. Genome screen of late-onset Alzheimer’s extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:50–55. | ||
Evans DA, Bennett DA, Wilson RS, et al. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003;60:185–189. | ||
Tang MX, Maestre G, Tsai WY, et al. Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet. 1996;58:574–584. | ||
Irie F, Fitzpatrick AL, Lopez OL, et al. Enhanced risk for Alzheimer disease in persons with type 2 diabetes and APOE epsilon4: the cardiovascular health study cognition study. Arch Neurol. 2008;65:89–93. | ||
Murrell JR, Price B, Lane KA, et al. Association of apolipoprotein E genotype and Alzheimer disease in African Americans. Arch Neurol. 2006;63:431–434. | ||
Myers RH, Schaefer EJ, Wilson PW, et al. Apolipoprotein E epsilon4 association with dementia in a population-based study: the Framingham study. Neurology. 1996;46:673–677. | ||
Fabian VA, Jones TM, Wilton SD, et al. Alzheimer’s disease and apolipoprotein E genotype in Western Australia: an autopsy-verified series. Med J Aust. 1996;165:77–80. | ||
Yu JT, Tan L, Hardy J. Apolipoprotein E in Alzheimer’s disease: an update. Annu Rev Neurosci. 2014;37:79–100. | ||
Raygani AV, Zahrai M, Raygani AV, et al. Association between apolipoprotein E polymorphism and Alzheimer disease in Tehran, Iran. Neurosci Lett. 2005;375:1–6. | ||
Bahia VS, Kok F, Marie SN, et al. Polymorphisms of APOE and LRP genes in Brazilian individuals with Alzheimer disease. Alzheimer Dis Assoc Disord. 2008;22:61–65. | ||
Qiu C, Kivipelto M, Aguero-Torres H, Winblad B, Fratiglioni L. Risk and protective effects of the APOE gene towards Alzheimer’s disease in the Kungsholmen project: variation by age and sex. J Neurol Neurosurg Psychiatry. 2004;75:828–833. | ||
Kardaun JW, White L, Resnick HE, et al. Genotypes and phenotypes for apolipoprotein E and Alzheimer disease in the Honolulu-Asia aging study. Clin Chem. 2000;46:1548–1554. | ||
Bickeboller H, Campion D, Brice A, et al. Apolipoprotein E and Alzheimer disease: genotype-specific risks by age and sex. Am J Hum Genet. 1997;60:439–446. | ||
Ganguli M, Chandra V, Kamboh MI, et al. Apolipoprotein E polymorphism and Alzheimer disease: the Indo-US Cross-national dementia study. Arch Neurol. 2000;57:824–830. | ||
Davidson Y, Gibbons L, Pritchard A, et al. Apolipoprotein E epsilon4 allele frequency and age at onset of Alzheimer’s disease. Dement Geriatr Cogn Disord. 2007;23:60–66. | ||
Molero AE, Pino-Ramirez G, Maestre GE. Modulation by age and gender of risk for Alzheimer’s disease and vascular dementia associated with the apolipoprotein E-epsilon4 allele in Latin Americans: findings from the Maracaibo Aging Study. Neurosci Lett. 2001;307:5–8. | ||
Payami H, Montee KR, Kaye JA, et al. Alzheimer’s disease, apolipoprotein E4, and gender. JAMA. 1994;271:1316–1317. | ||
Anttila T, Helkala EL, Kivipelto M, et al. Midlife income, occupation, APOE status, and dementia: a population-based study. Neurology. 2002;59:887–893. | ||
Slooter AJ, Cruts M, Kalmijn S, et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam study. Arch Neurol. 1998;55(7):964–968. | ||
Miech RA, Breitner JC, Zandi PP, et al. Incidence of AD may decline in the early 90s for men, later for women: the Cache County study. Neurology. 2002;58(2):209–218. |
Supplementary material
  | Table S1 Hardy–Weinberg equilibrium of APOE in AD patients and controls |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.