Back to Journals » International Journal of Nanomedicine » Volume 15
Antiviral Activity of Chitosan Nanoparticles Encapsulating Curcumin Against Hepatitis C Virus Genotype 4a in Human Hepatoma Cell Lines
Authors Loutfy SA , Elberry MH , Farroh KY, Mohamed HT , Mohamed AA, Mohamed EB, Faraag AHI , Mousa SA
Received 10 December 2019
Accepted for publication 23 March 2020
Published 22 April 2020 Volume 2020:15 Pages 2699—2715
DOI https://doi.org/10.2147/IJN.S241702
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
This paper has been retracted
Samah A Loutfy, 1, 2 Mostafa H Elberry, 1 Khaled Yehia Farroh, 3 Hossam Taha Mohamed, 4, 5 Aya A Mohamed, 4 ElChaimaa B Mohamed, 1 Ahmed Hassan Ibrahim Faraag, 6 Shaker A Mousa 7
1Virology and Immunology Unit, Cancer Biology Department, National Cancer Institute, Cairo University, Cairo, Egypt; 2Nanotechnology Research Center, British University, Cairo, Egypt; 3Nanotechnology and Advanced Materials Central Lab, Agricultural Research Center, Giza, Egypt; 4Faculty of Biotechnology, October University for Modern Sciences and Arts, 6th October, Giza, Egypt; 5Department of Zoology, Faculty of Science,Cairo University, Giza, Egypt; 6Botany and Microbiology Department, Bioinformatics Center, Faculty of Science, Helwan University, Cairo, Egypt; 7The Pharmaceutical Research Institute, Albany College of Pharmacy and Health Sciences, Rensselaer, NY, USA
Correspondence: Samah A Loutfy
Virology and Immunology Unit, Cancer Biology Department, National Cancer Institute, Cairo University, Kasr El Eini, Fom El-Khalig, Cairo 11796, Egypt
Tel +20 1222840964
Email [email protected]
Purpose: Current direct-acting antiviral agents for treatment of hepatitis C virus genotype 4a (HCV-4a) have been reported to cause adverse effects, and therefore less toxic antivirals are needed. This study investigated the role of curcumin chitosan (CuCs) nanocomposite as a potential anti-HCV-4a agent in human hepatoma cells Huh7.
Methods: Docking of curcumin and CuCs nanocomposite and binding energy calculations were carried out. Chitosan nanoparticles (CsNPs) and CuCs nanocomposite were prepared with an ionic gelation method and characterized with TEM, zeta size and potential, and HPLC to calculate encapsulation efficiency. Cytotoxicity studies were performed on Huh7 cells using MTT assay and confirmed with cellular and molecular assays. Anti-HCV-4a activity was determined using real-time PCR and Western blot.
Results: The strength of binding interactions between protein ligand complexes gave scores with NS3 protease, NS5A polymerase, and NS5B polymerase of -124.91, -159.02, and -129.16, for curcumin respectively, and -68.51, -54.52, and -157.63 for CuCs nanocomposite, respectively. CuCs nanocomposite was prepared at sizes 29– 39.5 nm and charges of 33 mV. HPLC detected 4% of curcumin encapsulated into CsNPs. IC50 was 8 μg/mL for curcumin and 25 μg/mL for the nanocomposite on Huh7 but was 25.8 μg/mL and 34 μg/mL on WISH cells. CsNPs had no cytotoxic effect on tested cell lines. Apoptotic genes’ expression revealed the caspase-dependent pathway mechanism. CsNPs and CuCs nanocomposite demonstrated 100% inhibition of viral entry and replication, which was confirmed with HCV core protein expression.
Conclusion: CuCs nanocomposite inhibited HCV-4a entry and replication compared to curcumin alone, suggesting its potential role as an effective therapeutic agent.
Keywords: caspase-dependent pathway, chitosan curcumin nanocomposite, hepatitis C virus genotype 4a, Huh7, docking
Corrigendum for this paper has been published
Plain Language Summary
Hepatitis C virus genotype 4a (HCV-4a) is a major public health problem, especially among the Egyptian population. There are more than 70 million individuals infected worldwide. It is the leading cause of chronic liver diseases, cirrhosis, and hepatocellular carcinoma. Newly emerged anti-HCV viral agents have recently become available on the market but with serious complications, high cost, and possible development of resistance. This has encouraged scientists to search for alternative, safer antiviral approaches. Nanoparticles offer unique physical properties that have associated benefits for antimicrobial activity or as drug carrier that can improve antiviral therapy. The significance of our research is in establishment of a natural nanoparticle drug carrier system that we screened with computer simulation studies and in cells against HCV-4a replication into human hepatoblastoma cell line. This may serve as a model for the screening of newly developed antiviral agents targeting HCV-4a to help in controlling this chronic disease.
Introduction
Hepatitis C virus (HCV) currently infects over 71 million individuals around the world. Its persistence causes infected individuals to enter a chronic phase that then leads to several different outcomes of liver disease.1
HCV therapies are becoming highly efficient and well tolerated due to improved understanding of HCV biology and to the discovery of antiviral targeting of essential viral functions.1,2 The NS5B polymerase, NS5A and NS3 proteases are considered crucial targets for the development of direct-acting antiviral drugs.3
The current treatment of HCV genotype 4a (HCV-4a) includes sofosbuvir, ledipasvir, and ombitasvir. The response rate has reached more than 90%.4 Despite the promise of such therapy, the suboptimal efficacy and adverse effects like anemia, rash, bilirubin, nausea, pruritus, and photosensitivity5 as well as the increasing number of patients with advanced liver disease and no therapeutic options emphasize a major need for improved therapeutic regimens. In addition, the high mutation rate of viral RNA and development of mutant resistant strains has slowed development of innovative anti-HCV drugs. Therefore, natural compounds extracted from plants have attracted the attention of researchers for their therapeutic activities.6 A substantial variety of natural compounds has demonstrated antiviral activity, including against HCV,7 or hepatoprotective effect from natural compounds like naringenin, epigallocatechin-3-gallate (EGCG), silymarin, and caffeine.8,9
Turmeric curcumin, known as curcuma longa, is a natural plant with a significant therapeutic role that harbors no side effects.10 Curcumin’s activity as an antiviral emerged recently, and it exhibits a significantly high inhibition against several viruses, like hepatitis B virus,11 HIV, influenza viruses, and human herpes viruses.12 Several reports demonstrated curcumin’s activity against HCV; it can obstruct viral attachment and fusion to hepatocytes and prevent cell to cell transmission due to loss of the membrane’s integrity.10,13
However, curcumin’s positive effect is hampered by its lower solubility, bioavailability, and low cellular uptake, but the progress in nanotechnology has enabled formulation of nanoparticles, and polymeric nanoparticles can be used to encapsulate curcumin. This could resolve curcumin’s limitations by enhancing its sustained release to target diseased cells, improve its bioavailability, prevent it from degradation or metabolism, and increase its therapeutic potential.14 Polymeric nanoparticles are biocompatible and biodegradable and have been developed as drug delivery vectors and have desirable properties like better encapsulation of compounds to protect and deliver them efficiently with high pharmacokinetics and endocytosis efficiency.15
Chitosan is derived from chitin, which is a natural biopolymer and is the second most abundant natural polysaccharide.16 It is found primarily in marine animals’ exoskeleton, ie, lobsters, shrimps and crabs. Chitosan is immunogenic, and its antitumor, anti-inflammatory, antibacterial, antifungal, and antimicrobial activities have been observed.17,18 It has been reported that preparation of chitosan as nanoparticles revealed no toxicity at lower concentration (30 µg/mL) but showed anticancer activity at higher concentrations (>100 µg/mL) against different cancer cell lines, which supports its application as a drug delivery system.17,19
Previously, chitosan nanoparticles (CsNPs) have been conjugated with antiretroviral agents like saquinavir, a protease inhibitor, to improve anti-HIV therapy; cell targeting efficiency increased by 92% compared to the soluble drug control.20 Accordingly, to increase the antiviral activity of curcumin, in the present study we conjugated chitosan and curcumin as a nanocomposite and evaluated it for antiviral activity against entry and replication of HCV-4 in hepatoblastoma cells and performed in silico studies.
Materials and Methods
Materials
Curcumin and Bradford protein assay kits were purchased from Bio Basic Inc. (Markham, ON, Canada). Phosphate-buffered saline (PBS) tablets were purchased from Fisher Scientific (Loughborough, UK). Chitosan (low molecular weight), tripolyphosphate (TPP), ethanol, methanol, ethyl acetate, glutaraldehyde, and isopropanol were purchased from Sigma‐Aldrich (St Louis, MO, USA). MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reagent was purchased from Serva (Heidelberg, Germany). Dulbecco’s modified Eagle’s medium (DMEM), penicillin, streptomycin, and fetal bovine serum (FBS) were purchased from Gibco (Merelbeke, Belgium). Paclitaxel was purchased from Actavis Pharma, Inc. (Parsippany, NJ, USA). The Huh7 cells were supplied by Rockefeller University (New York, NY, USA). The WISH cell line was obtained from Vaccination and Sera Collection Organization (VACSERA, Agouza, Giza, Egypt).
In silico Studies
In silico docking was performed using Molegro Virtual Docker (MVD) 2013.6.0.0 (CLC bio-Qiagen Company, Aarhus, Denmark).21 The 3D structure of CuCs nanocomposite was predicted using Schrödinger’s Materials Science Suite 2.6 (Figure 1). Curcumin ligand was retrieved from PubChem Bioassay. Homology models of NS3 protease and NS5A polymerase were built using the prime program of the Schrödinger software suite. Blast search was performed to identify a template protein structure.22 Loops refinement and verification was performed using the protein refinement program of Schrödinger software.23–25 The binding sites of NS3 protease, NS5A polymerase, and NS5B polymerase were detected using MVD cavities prediction. All docking calculations were carried out using the grid-based MolDock score (GRID) function with 0.30 Å grid resolution.
Preparation of CsNPs and CuCs Nanocomposite
The CsNPs were prepared using an ionotropic gelation method of chitosan with TPP anions where positive charges of the amino groups of chitosan were slightly modified to be crosslinked with the TPP negative charges as we presented previously.19 CuCs nanocomposite was prepared by dissolving 320 mg of curcumin in 80 mL ethanol and incubated with 500 mg CsNPs suspended in 40 mL of Millipore water at acidic pH from 4.0 to 5.0 with some modifications.17,26 Finally, the curcumin was bound to CsNPs and dried using vacuum.
Characterization of CsNPs and CuCs Nanocomposite
Transmission Electron Microscopy (TEM)
High-resolution TEM was used to determine the size, shape, and size distribution of the CsNPs and CuCs nanocomposite by using a drop from diluted nanoparticles solution in order to form a monolayer through deposition on an amorphous carbon-coated copper grid and left to evaporate at room temperature. The particle and size distribution measurements were identified with a JEM-2100 transmission electron microscope (JEOL, Tokyo, Japan) that accelerates voltage at 200 kV, and the imaging was performed using the same microscope.
Zeta Potential Measurement
Zeta potential was performed to measure the electrical charges of the particles in nano colloids using a Nano ZS apparatus (Malvern Instruments, Malvern, UK), through electrophoretic light scattering of the nanoparticles in an aqueous solution and using Smoluchowski equation to calculate the zeta potentials at 25ºC.27 The measurement was completed during 6 minutes and the migration voltage was fixed at 25 V.
Fourier Transform Infrared Spectroscopy (FTIR)
The FTIR was carried out for CsNPs, curcumin, and CuCs nanocomposite to show at which bonds the curcumin and chitosan were formed. FTIR was used with the following specifications: spectral range 400–4500 cm−1, number of scans 16, spectral resolution 4 cm−1. The software was Opus (Bruker (Germany), VERTEX 70, RAMII).
High-Performance Liquid Chromatography (HPLC)
Nanoparticles from the nanocomposite were weighed and acidified at pH 3.0; curcumin was extracted from the compound using a mixture of ethyl acetate and isopropanol (9:1 v/v) by shaking for 6 minutes. The mixture was centrifuged at 3864 × g for 20 minutes, and the upper layer of ethyl acetate was discarded and the extraction step was repeated. The final extraction solution was evaporated with vacuum to be completely dry, and its residue was dissolved using 100 μL ethanol. An aliquot of the dissolved solution was used for curcumin quantification via HPLC by calculating the peak area of absorbance of the samples at wavelength 428 nm and comparing it to the standard peak area of curcumin. Finally, the mobile phase was prepared by mixing 1% citric acid pH 3.0/acetonitrile (45:55, v/v), and the flow rate was 1 mL/min.
In vitro Drug Release
The release of curcumin from its encapsulation in CsNPs was performed at pH 7.4, and the nanoparticles were re-dispersed in PBS. The total volume of the solution was divided into 5 tubes at 37°C under orbital shaking. Subsequently, curcumin in nanoparticles was centrifuged at 966 × g for 10 minutes, where the sediment was extracted in methanol and quantified using spectrophotometry. Finally, the release of curcumin from CsNPs was quantified according to the following equation:
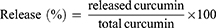
Cell Culture
Huh7 cells, derived from human hepatoma cells, were received as a gift from the Lab of Radiobiology and Experimental Radiooncology, UKE, Hamburg, Germany and were used and maintained at Virology and Immunology unit, Cancer Biology Dept., National Cancer Institute, Cairo University. The WISH cell line, human amniotic cells, was used as a model for normal cells. The cells were maintained as a monolayer in a 25 cm2 flask with approximately 6 mL DMEM supplemented with 10% FBS, 2% penicillin, and 2 mg/mL streptomycin. The cells were incubated under standard conditions of 37ºC, 5% CO2, and 95% humidity.
Cytotoxicity and MTT Colorimetric Assay
Cellular toxicity of the tested materials (curcumin, CsNPs and CuCs nanocomposite) was investigated against Huh7 cells using 2-fold dilutions starting from 100 to 6.25 µg/mL, according to our previously published protocol.19 The MTT formazan product was identified via measuring the absorbance using an enzyme linked immunosorbent assay (ELISA) plate reader (Model ELX800, BioTek Instruments, Inc., Winooski, VT, USA), and positive and negative controls were run in the plate. The viability of cells (%) in relation to the control wells with untreated cells was calculated using the following equation:
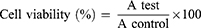
where A test is the absorbance of the test sample and A control is the absorbance of the control sample. The results were the average of three wells, and 100% viability was determined from the negative control (untreated cells).
Morphological Investigation
The cytotoxic effect of the IC50 concentration of CuCs nanocomposite was followed microscopically with phase contrast microscopy (100x magnifications) after treatment of Huh7 and Wish cells after 24 hours.
Combination Index and Fraction Effect
Compusyn software (version 1.0, ComboSyn, Inc., Paramus, NJ, USA) was used to predict and simulate the combination index (CI) and fraction effect (fa) of the CuCs nanocomposite to estimate its activity and the amount of drug released into cells.28 Simulated CuCs nanocomposite was used to investigate its cellular response using a response additivity model (Linear Interaction Effect). Such a model can demonstrate the positive effect of a drug combination (chitosan and curcumin) that might occur when the combination effect (EAB) is greater than the expected additive effect given by the sum of the individual effects (EA+EB). Therefore, we investigated such possible favorable outcomes for better efficacy of therapeutic application and the minimal development of drug toxicity to provide a selective synergistic effect against the target. Accordingly, this can be achieved by using a minimal set of mathematical and pharmacological concepts, according to an equation where the CI was calculated as follows: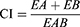
where EA refers to curcumin, EB refers to CsNPs and EAB refers to the nanocomposite. The resulting CI theorem offers a quantitative definition for additive effect (CI = 1), synergism (CI < 1), and antagonism (CI > 1) in drug combinations. Such analysis was also performed to confirm the toxic effect of curcumin when combined with CsNPs, which also may indicate its possible role in the antiviral mechanism.
Cellular Uptake of Curcumin Chitosan Nanocomposite
The in vitro cellular uptake of the particles was carried out using TEM, and the cells were first counted using a hemocytometer. Then, the cells (3 × 106/75 cm2 flask) were treated with 100 µM of CuCs nanocomposite for 24 hours. The cells were then washed with PBS buffer, fixed with 2% glutaraldehyde for 2 hours, then washed twice with PBS and finally fixed in 1% OsO4 for 1 hour. Following agarose (1.5%) enrobing, Spurr’s resin embedding, and ultrathin (50 nm) sectioning, the samples were stained with 2% aqueous uranyl acetate and 25 mg/mL lead citrate and imaged with a JEOL 100S microscope,29 but cell control was tested using a field emission scanning electron microscope equipped with a scanning transmission electron (STEM) detector (Quattro S, Thermo Fisher Scientific).
Flow Cytometry Analysis of Curcumin Chitosan Nanocomposite
Flow cytometry was performed to identify the nature of cell death that was induced after treatment of Huh7 with IC50 concentration of CuCs nanocomposite. This was done by trypsinization of cells and centrifugation at 314 × g for 5 minutes. Then, 100 µL of cell suspension was added to 100 µL of DNA PREP LPR (<0.1% potassium cyanide, <0.1% NaN3, nonionic detergents, saline, and stabilizers) to obtain final cell concentration of 3–5 x106 cells/mL. Afterward, 2 mL of DNA PREP stain was added to the cell suspension solution and incubated at room temperature for 30 minutes. Finally, acquisition was done on a flow cytometer (Beckman Coulter, EpicsXL, Brea, CA, USA) of 10,000 events to calculate the percentages of cells occupying the different phases of the cell cycle.30
Quantification of mRNA Expression of Apoptotic Genes; Caspase 3 and Bcl-2
Two 25 cm2 flasks were seeded with 3 × 105 Huh7cells and were left for 24 hours under optimum culture conditions to have a confluent sheet of cells, and then cells were treated with the estimated IC50 concentration of curcumin and CuCs nanocomposite. Afterwards, cells were harvested and total RNA extraction was performed using nucleic acid extraction kit NucleoSpin® (Macherey-Nagel GmbH, Duren, Germany) according to the manufacturer’s instructions. The RNA extract was used in real-time PCR assay using SensiFAST™ SYBR® Hi-ROX One-Step Kit (Bioline, London, UK) with a total reaction volume of 25 µL and primers specific for caspase 3, Bcl-2 and the housekeeping gene β-actin. The reaction consisted of three stages: stage 1 at 45°C for 10 minutes, stage 2 at 95°C for 2 minutes, stage 3 repeated for 35 cycles that consisted of 95°C for 5 seconds, then 60°C for 10 seconds, and 72°C for 5 seconds. Finally, calculation of the relative quantification of the cycle threshold (Ct) was conducted according to the reaction of each target gene. The relative quantification (RQ) of each gene was quantified according to the calculation of ΔΔCt as the RQ of each gene is calculated by taking its 2−∆∆Ct according to the following equation:
ΔΔCt = [(Ct target, Sample)-(Ct ref, Sample)]-[(Ct target, Control)-(Ct ref, Control)]
where (Ct target, Sample) = Ct value of gene of interest in tested sample; (Ct ref, Sample) = Ct value of reference gene in tested sample; (Ct target, Control) = Ct value of gene of interest in control DNA; (Ct ref, Control) = Ct value of reference gene in control DNA.31
Protein Expression of Caspase 3 and Bcl-2
Six well tissue culture plates were seeded with 3 × 104 of Huh7 cells and left for 24 hours under optimum culture conditions to have a confluent sheet of cells, and then they were treated with the estimated IC50 concentration of the CuCs nanocomposite and paclitaxel as a standard chemotherapeutic agent to allow its comparison with nanocomposite. Afterwards, the cells were harvested and their total proteins were extracted to measure the levels of apoptotic proteins caspase 3, caspase 8, and p53, as well as levels of the anti-apoptotic marker Bcl-2. Then, the isolated proteins’ levels were measured using human Bcl-2 platinum ELISA assay kit according to the manufacturers’ instructions (eBioscience, Madison, WI, USA). The standard curve for each protein was drawn according to each kit and the reaction products were measured at 450 nm using a microplate reader (Tecan Group Ltd., Seestrasse, Männedorf, Switzerland).
Antiviral Assay
Six well tissue culture plates were seeded with 500,000 cells/well; Huh7cells were treated with nontoxic doses of curcumin, CsNPs, and CuCs nanocomposite at concentrations of 15 μg/mL, 100 μg/mL, and 20 μg/mL, respectively. This was performed according to a previously published protocol.32 A 500 µL aliquot of the positive serum for HCV-4a was inoculated and kept for 90 minutes for adsorption. DMEM (10%) was added to a final volume of 3 mL and the incubation was continued for 24 hours. Infected cells were treated with either curcumin, chitosan, or CuCs nanocomposite. Untreated infected cells and cell control were treated with media only. The plates were incubated for 24 hours, and viral RNA was extracted prior to real-time PCR assay according to the previously published protocol.32
Quantification of HCV Core Protein Using Western Blot
The six well plates were again used as previously described in the antiviral assay, then the infected and treated Huh7 cells were subjected to cell harvesting with a cell scraper, and the protein content of the cells was extracted using lysis buffer, then the protein content was measured with a Bradford protein assay kit. Subsequently, 20 µg of protein concentration of each sample was loaded into 10% SDS-PAGE with an equal volume of 2x Laemmli sample buffer (4% SDS, 10% β-mercaptoethanol, 2% glycerol, 0.004% bromophenol blue, and 0.125 M tris HCL). Afterward, the gel was transferred to a polyvinylidene fluoride (PVDF) membrane and then blocked with tris-buffered saline with Tween 20 buffer and 3% bovine serum albumin (BSA) for 1 hour. The membrane was incubated overnight at 4°C with 1:1000 dilution of the primary antibody (anti-HCV core 1b antibody, ab2740, Thermo Scientific, Boston, MA, USA) and then incubated for 1 hour at room temperature with horseradish peroxidase HRP-conjugated secondary antibody goat anti-rabbit IgG. Finally, the membrane was washed and bands were visualized with the aid of chemiluminescent substrate, and signals were detected using a CCD camera-based imager. Ultimately, band images of the target proteins were analyzed using ChemiDoc MP Imager in relation to β-actin as a positive control.33
Results
The workflow of methods performed in the current research is presented graphically in Scheme 1.
 |
Scheme 1 An illustration of the workflow of methods performed in the current research. Abbreviation: CuCs, curcumin chitosan. |
In silico Studies
The in silico study showed that the binding affinity of curcumin to NS3 protease and NS5A polymerase was higher than that of CuCs nanocomposite chitosan. However, CuCs nanocomposite showed good binding affinity toward NS5B polymerase as compared to curcumin. Curcumin showed a higher docking score toward NS3 protease and NS5A polymerase with −124.91 and −159.02 MolDock score, respectively (Table 1).
Binding of curcumin with HCV-4a target proteins (NS3, NS5A and NS5B) showed that it could form H-bonds at the cavity of NS3 protease (Figure 1A and B and Table 1) and NS5A polymerase (Figure 1C and D and Table 1), where it forms 2 H-bonds and 5 H-bonds with NS3 protease and NS5A polymerase inside the protein cavity, respectively. However, CuCs nanocomposite formed 9 H-bonds with NS5B polymerase (Figure 1E and F and Table 1). Curcumin formed one H-bond with Gly521 (B) and Leu 522 (B) of NS3 protease active site, despite the binding affinity of interaction toward NS5A being much higher than NS3 because it forms 5 H-bonds, one with Arg 160 (B), Leu 149 (A) and Gly96 (B) and two H-bonds with Trp111 (A). Interaction of CuCs nanocomposite in the case of NS5B polymerase is more effective because it forms 9 H-bonds, one with Arg 89, Arg 394, Cys 39, Gul 18, Pro 16, Ser 269 and Tyr 162 and two H-bonds with Arg 401.
Characterization of CuCs Nanocomposite
CuCs nanocomposite was subjected to several characterization procedures to study encapsulation efficiency. The TEM results showed that the size ranged from 29 to 39.5 nm (Figure 2A) with charges of 33 mV as measured with the zeta analyzer (Figure 2B). The FTIR spectra of chitosan, curcumin, and CuCs nanocomposite are shown in Figure 2C. Characteristic bands at 1511 cm−1 and 1062 cm−1 were attributed to the stretching vibrations of the C=C vibrations, and C-O-C stretching modes, respectively, in curcumin. For CsNPs, the characteristic band at 3450 cm−1 was assigned to stretching vibration mode of N-H overlapped with O-H stretching vibration mode, the band at 2880 cm−1 was due to C-H stretching vibration mode, the band at 1653 cm−1 was assigned to the C=O, and the band at 1428 cm−1 was assigned to bending vibration of N-H stretching vibration. In comparison with curcumin, a different spectrum was observed for CuCs nanocomposite as new bands appeared at 1070 cm−1 and the N-H group of CsNPs disappeared (Table 2). It can be assumed that the ammonium groups of chitosan were linked with OH groups of curcumin to form the nanocomposite. Results from HPLC indicate that 42 mg curcumin was encapsulated in the CsNPs, giving 4% of curcumin encapsulated inside CsNPs (Figure 2D and E).
 |
Table 2 Position of Bands in FTIR Spectra of Curcumin, Chitosan, and Curcumin Chitosan (CuCs) Nanocomposite |
The in vitro drug release experiment showed that initially, a gradual increase with the exponential release rate of curcumin was observed for the first 2 hours with an increment rate of 30 (Figure 2F). This large increase is due to the curcumin molecules that deposited on the surface of Cs nanocapsules. Afterward, there was a relatively constant release of curcumin for the next 4 hours and there was a linear plateau in release that was observed with a rate of −1.89 × 10−1. Supplementary Figure S1 shows the behavior profile of curcumin-encapsulated into CsNPs and the equation used for the calculation. From the release profile of curcumin, it is clear that about 45% of curcumin was released from the nanoparticles at time 0, and that afterward the amount of curcumin released from the nanoparticles was about 84% after the first 2 hours. Finally, there was no further release obtained for the other 4 hours.
Cell Culture
The cytotoxic effect of various concentrations of curcumin, chitosan, and CuCs nanocomposite showed that the IC50s of curcumin and CuCs nanocomposite were detected at concentrations of 8 and 25 µg/mL, respectively, on Huh7 after 48 hours of cell exposure, and CsNPs were nontoxic at more than 100 µg/mL on Huh7 or WISH (Figure 3). Regarding cytotoxic effects of the same materials on WISH, our results revealed that the IC50s of curcumin and CuCs nanocomposite were detected at concentrations of 25.8 and 34 µg/mL, respectively, after the same time of cell exposure.
 |
Figure 3 (A). IC50 of curcumin on Huh7, (B). IC50 of curcumin chitosan nanocomposite on Huh7, (C). IC50 of curcumin on WISH cells, (D). IC50 of curcumin chitosan nanocomposite on WISH cells. |
The light microscopy results showed aggressive changes at the concentration of the IC50 for both curcumin and CuCs nanocomposite without any changes in the morphology of cells treated with CsNPs at 100 µg/mL (Figure 4).
Calculation of Combination Index and Fraction Effect
The CuCs nanocomposite showed a good therapeutic effect against human liver cancer cells of fraction effect 75 that can kill 75% for the simulated drug release amount upon combining 48 µg/mL of curcumin and 193 µg/mL chitosan. But upon increasing these concentrations, the effect would be antagonistic according to CompuSyn software (Table 3, Supplementary Tables S1 and S2).
 |
Table 3 Combination Index (CI) of Curcumin, Chitosan Nanoparticles (CsNPs), and Curcumin Chitosan (CuCs) Nanocomposite on Huh7 Cells and Its Type of Interaction as Simulated with CompuSyn Software |
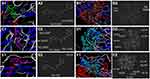
Cellular Analysis of Chitosan Nanoparticles and Curcumin Chitosan Nanocomposite in Huh7
TEM images showed binding and internalization of CsNPs into Huh7 cells. Aggregation of CsNPs to form nanoparticle clusters on the cell membrane and its dispersion in the cytoplasm was evident (Figure 5A–C).
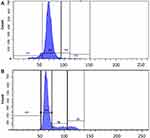
The CuCs nanocomposite was further investigated with flow cytometry of cell cycle and DNA contents of Huh7 cells treated with the IC50 concentration. Results showed cell arrest in the preG1 phase, preventing cells from entry into S and G2/M phases of the cell cycle (Figure 6A and B and Table 4), whereas the untreated control cells showed the expected cell cycle pattern for continuously growing cells.
 |
Table 4 Flow Cytometric Cell Cycle Analysis of Treated Huh7 Cells with IC50 Concentration of Curcumin Chitosan (CuCs) Nanocomposite |
Quantification of Caspase 3 mRNA Apoptotic Gene and Anti-Apoptotic Bcl-2
The mRNA expressions of caspase 3 gene increased post-treatment with IC50 concentrations of curcumin, and it was doubled after treatment of cells with CuCs nanocomposite. However, the expression of Bcl-2 decreased post-treatment with both materials (Table 5, Supplementary Figure S2).
 |
Table 5 Quantitative Real-Time PCR Assay for Caspase 3, Bcl-2 Apoptotic Genes Relative to β-Actin as a Housekeeping Gene |
Effect of Treatment of Curcumin Chitosan Nanocomposite on Caspase 3, Caspase 8, P53 and Bcl-2 Protein Levels
Treatment of Huh7 cells with the IC50 concentrations of CuCs nanocomposite was associated with an increase in the activity of caspase 3 protein from 9.7 µg/mL to 12 µg/mL, and it markedly decreased the protein level of anti-apoptotic Bcl-2 from 2.8 µg/mL to 0.2 µg/mL. Moreover, a marked increase in the protein levels of caspase 8 and p53 is shown in Figure 7A. Likewise, the effect of paclitaxel as a standard anticancer drug on treatment of Huh7 cells with IC50 concentration of 2 µM paclitaxel resulted in a noticeable increase in levels of caspase 3 and marked decrease in Bcl-2, as well as a noticeable increase in protein levels of caspase 8 and p53 (Figure 7B).
Antiviral Assay Against Viral Entry in Huh7 Cells
Antiviral activities of curcumin, CsNPs and CuCs nanocomposite were tested against the viral entry in infected Huh7 cells from positive HCV-4a patients through mixing of equal volumes of the nontoxic concentrations of the investigated materials and viral titer; in this assay one viral titer was involved (105 IU/mL). As a result, the tested compounds showed high antiviral activities against entry of HCV-4a into Huh7 cells by almost 100% reduction in viral titer, and curcumin inhibited viral entry by almost 95%. (Table 6).
 |
Table 6 Antiviral Activity of Nontoxic Doses of Curcumin, Chitosan Nanoparticles (CsNPs), and Curcumin Chitosan (CuCs) Nanocomposite Against Entry of HCV 4 into Huh 7 Cells |
Quantification of HCV Core Protein
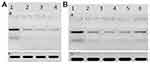
The HCV core protein expression level post-treatment of HCV infected Huh7 cells with curcumin, CsNPs, and CuCs nanocomposite against viral replication and entry was quantified in relation to β–actin as a control (Table 7, Figure 8). The expression post-treatment against viral replication showed a decrease in cells treated with the three compounds and specifically for that treated with nanocomposite with a range 0.9 to 5.6 ratio to the positive infected Huh7 cells. Regarding expression post-treatment with curcumin, CsNPs, and nanocomposite against viral entry, there was a decrease in the HCV core protein expression, especially in those treated with nanocomposite with a 1.05 to 5.6 ratio to the positive infected Huh7 cells.
 |
Table 7 HCV Core Protein Expression Post Treatment of Huh7 Analyzed with Western Blot Assay with Curcumin, Chitosan Nanoparticles (CsNPs), and Curcumin Chitosan (CuCs) Nanocomposite |
Discussion
Recently, the advent of new direct-acting antiviral drug therapy for HCV-4a has greatly improved sustained virology response, although there is the possibility of serious adverse effects and development of resistant mutant strains. Therefore, discovery of more effective and less toxic antiviral agents is still needed. Computational screening of large chemical libraries using molecular docking and the establishment of HCV culture model has led to numerous studies and the discovery of many anti-HCV agents. Accordingly, we started working on CsNPs,19 and the in silico results demonstrated high binding affinity towards HCV-4a target proteins (NS3 protease, NS5B polymerase, and NS5A helicase) and this encouraged us to continue our research on CsNPs as a drug delivery system for antiviral agents, hoping to increase therapeutic efficacy and decrease therapeutic dose. In the current research, curcumin was encapsulated into CsNPs and evaluated for its antiviral activity against HCV-4a first with in silico analysis and subsequently with in vitro studies, with the hope to discover new candidates for use as HCV-4a inhibitors. Our in silico analysis gave promising results by showing an increase in the affinity of our designed nanocomposite against HCV-4a NS5B compared to curcumin alone, suggesting its potential role against viral replication, first with in silico analysis and subsequently with in vitro studies, to discover new candidates for use as HCV-4a inhibitors. Our in silico analysis gave promising results by showing an increase in the affinity of our designed nanocomposite against HCV-4a NS5B compared to curcumin alone, suggesting its potential role against viral replication.
Here we found that curcumin conjugated to chitosan via the OH group of curcumin and the ammonia group of CsNPs crosslinked with TPP, which resulted in small sizes (29.6–39.5nm)34 compared to results previously published by Das and colleagues who used sodium alginate as a crosslinker instead of TPP salt, which led to chitosan with a bigger size (100±20 nm).35 Different fundamental and clinical investigations illustrate curcumin’s restricted adequacy because of its poor dissolvability, rapid metabolism, low bioavailability, and pharmacokinetics. In addition, its physicochemical properties are unstable in several conditions like neutral pH or alkaline pH ≥7.0, and ambient temperature.36 Therefore, we conjugated curcumin into CsNPs to improve its properties, control its release, and enhance its antiviral activity. Our preparation was performed at acidic pH with certain modifications,26 using ascorbic acid as solvent for CsNPs instead of acetic acid, where positive charges of amino groups on chitosan can interact with negative charges on OH of curcumin through electrostatic interactions, leading to formation of CuCs nanocomposite. This binding property was found to be much less at neutral or alkaline pH due to the higher tendency of CsNPs to aggregate because of neutralization of the amino groups on CsNPs.17 Our HPLC experiment demonstrated 4% entrapment of curcumin into CsNPs after 6 minutes of polymeric digestion; this time needs to be lengthened in the future preparations. Drug release profile showed that 50% of loaded curcumin was released during the first 11 minutes—the release time known as “burst release” meaning the amount of curcumin deposited on the outer surface of the CsNPs—and 34% of curcumin was released at 120 minutes. This means that the total release of curcumin (84%) was obtained after 2 hours. Afterward, a relatively constant release of curcumin was observed for the next 4 hours (ie 240 minutes). This is supported by results of Akhtar et al who reported pharmacokinetic analysis for chitosan bound to curcumin in parasite Plasmodium yoelii infected mice.17 They found that the curcumin concentration detected in the blood was significantly higher in mice fed with curcumin loaded onto CsNPs compared to blood from mice administered an equimolar concentration of curcumin alone. Results showed that administration of CsNPs orally ensured a sustained release of curcumin over 8 hours post-feeding, whereas, in the case of curcumin alone, the levels declined significantly after 3 hours and were not detectable beyond 6 hours.
In a study by Samrot et al, curcumin-loaded CsNPs were synthesized from crab shell for drug delivery using two chelators (TPP and BaCl2) in preparation of CsNPs.37 They observed an initial drug burst from the biopolymer that then become steady, and the CsNPs chelated with BaCl2 led to sustained drug release for more than 70 minutes. Regarding CsNPs chelated with TPP, the samples had a stable drug release profile; drug release increased from the 20th to the 60th minute and then gradually decreased until the 100th minute, followed by a stable release. This indicates that chitosan synthesized using TPP showed a slow release of curcumin for 180 minutes.
Such an observation might explain why the cytotoxic effect of our synthesized nanocomposite was lower than that of curcumin alone (25 µg/mL versus 8 µg/mL for curcumin). The literature shows that the cytotoxic effect of a nanoformulation of curcumin is equal to or higher than that of free curcumin.38,39 This does not match our results where the cytotoxic effect of CuCs was lower than that of free curcumin. This might be due to i) the lower % of the entrapment efficiency, ii) the nature of the nanocarrier, which is different in the current study than in previous studies, and iii) the antioxidant properties of CsNPs as previously reported by Nair et al.40
Further extensive investigations using simulated CompuSyn software (Linear Interaction Effect) were conducted to investigate possible additivity between curcumin and CsNPs, which might predict the synergistic effect of CuCs nanocomposite as antiviral activity. Upon using multiple inhibitors of different types and mechanisms of inhibition on enzyme kinetic models, several hundred equations have been derived that can be reduced to general equation by comparing theoretical biological activities in the absence and presence of inhibitors. This mathematical work leads to the CI. The obtained data are unique to each laboratory because data vary according to instrument, reagents, and type of cells grown in each lab.41
The nature of cell death induced by the formulated CuCs nanocomposite was analyzed with flow cytometry, and results showed that Huh7 treated with an IC50 concentration of nanocomposite induced cell cycle arrest at the preG1 phase in comparison to the untreated cells, which hindered their entrance into the S and G2/M phases. This might be due to the small amount of DNA content and admission into programmed cell death, as previously illustrated by Kumar et al.42 The results of molecular apoptotic gene expression level of cells treated with an IC50 concentration of curcumin and its composite showed high expression levels in caspase 3 and lowered expression of Bcl2 in Huh7 cells treated with curcumin and its nanocomposite. This indicates that the apoptosis was induced via a caspase-dependent pathway. Such cytotoxic effect could be due to the production of reactive oxygen species in response to the toxic dose of curcumin, resulting in DNA breakage, which in turn causes activation of p53 that can recruit other molecules like Bax, Bak. This, in turn, causes pores in the mitochondria membrane, disrupting its membrane, leading to the release of cytochrome c, which stimulates a cascade of death compounds and ultimately cell death.43 Finally, protein expression levels of caspase 3, caspase 8, p53, and Bcl2 were detected using ELISA assay. Cells treated with paclitaxel, a standard anticancer drug served as a control. Our results matched that of mRNA transcriptional level, which showed an increase in the expression levels of caspase 3, caspase 8, and p53 proteins and a decrease in the expression levels of Bcl2 protein compared to untreated Huh7. These results confirmed the designated roles of the nanocomposite in mediation in the apoptotic process through cell death receptors and induction of apoptosis through activation of intrinsic and extrinsic apoptotic pathways.44,45
Concerning assessment of the antiviral activity of CsNPs, curcumin, and its nanocomposite, curcumin demonstrated inhibition of HCV-4a entry by 94.5%, which is in line with results reported by Kusuma et al,46 where curcumin was confirmed to block the HCV entry pathway into its targeted hepatoma cells with no effect on viral assembly or virion release. Moreover, our data match those of Anggakusuma et al who reported that curcumin nanoformulation increased its antiviral activity with no toxic effect; they demonstrated that the antiviral mechanism occurred by affecting virion membrane fluidity without disrupting virion integrity.10 In the present study, the same observations were obtained upon analyzing antiviral activity against replication of HCV-4a; these results confirm the in silico analysis and demonstrate the dual roles of CsNPs and its composite to inhibit both viral entry and replication. These results were confirmed with HCV core protein expression using Western blot assay, which demonstrated a remarkable decrease in the expression level after the treatment of cells with CsNPs, curcumin, and nanocomposite. Nevertheless, the nanocomposite exhibited the lowest decrease in the HCV core protein expression, which indicates that the nanocomposite has the highest antiviral activity whether against viral entry or replication. Further investigations are still required to study the anti-HCV activity of nanocomposite in the replicating system, which is underway in our laboratory.
In conclusion, the current study obtained a clear view on the potency of CsNPs in synergizing the antiviral activity of curcumin on binding as a nanocomposite form. Additionally, the prepared nanocomposite demonstrated a powerful multitarget antiviral agent against HCV-4a entry and replication, and this was proved on the molecular and protein levels. However, it is mandatory to confirm the novelty of the nanocomposite as a potential alternative therapy against HCV-4a infection through analysis of pharmacokinetics and pharmacodynamics as well as further analysis on the HCV-4a replicating system to obtain the therapeutic index and ultimately an in vivo study.
Ethics and Consent Statement
The research has been approved by the National Cancer Institute-Cairo University, IRB IRB00004025 Organization No. IORG0003381.
Acknowledgments
This project was supported financially by the Science and Technology Development Fund (STDF), Egypt, Grant No.15108. We would like to thank National Cancer Institute, Cairo University, for supporting us with equipment, reagents, and consumables. Our sincere gratitude goes to the Lab of Radiobiology and Experimental Radiooncology, UKE, Hamburg, Germany, for supplying Huh7 and WISH cell lines as a gift to complete our research. We also thank Ahmed Mamdouh Osman for his technical assistance.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. World Health Organization. Hepatitis C; 2019. Available from: http://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
2. Elgharably A, Gomaa AI, Crossey MME, Norsworthy PJ, Waked I, Taylor-Robinson SD. Hepatitis C in Egypt – past, present, and future. Int J General Med. 2016;10:1–6. doi:10.2147/IJGM
3. Tremblay N, Young Park A, Lamarre D. HCV NS3/4A protease inhibitors and the road to effective direct-acting antiviral therapies. Hepatitis C Virus II. 2016. 257–285.
4. Boyd SD, Harrington P, Komatsu TE, et al. HCV genotype 4, 5 and 6: distribution of viral subtypes and sustained virologic response rates in clinical trials of approved direct-acting antiviral regimens. J Viral Hepat. 2018;25(8):969–975. doi:10.1111/jvh.2018.25.issue-8
5. Nafisi S, Roy S, Gish R, Manch R, Kohli A. Defining the possibilities: is short duration treatment of chronic hepatitis C genotype 1 with sofosbuvir-containing regimens likely to be as effective as current regimens? Expert Rev Anti Infect Ther. 2016;14(1):41–56. doi:10.1586/14787210.2016.1114883
6. Jardim ACG, Shimizu JF, Rahal P, Harris M. Plant-derived antivirals against hepatitis C virus infection. Virol J. 2018;15(1):34. doi:10.1186/s12985-018-0945-3
7. Calland N, Dubuisson J, Rouille Y, Seron K. Hepatitis C virus and natural compounds: a new antiviral approach? Viruses. 2012;4(10):2197–2217. doi:10.3390/v4102197
8. Assini JM, Mulvihill EE, Burke AC, et al. Naringenin prevents obesity, hepatic steatosis, and glucose intolerance in male mice independent of fibroblast growth factor 21. Endocrinology. 2015;156(6):2087–2102. doi:10.1210/en.2014-2003
9. Khalaf N, White D, Kanwal F, et al. Coffee and caffeine are associated with decreased risk of advanced hepatic fibrosis among patients with hepatitis C. Clin Gastroenterol Hepatol. 2015;13(8):1521. doi:10.1016/j.cgh.2015.01.030
10. Anggakusuma, Colpitts CC, Schang LM, Rachmawati H, et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2014;63(7):1137–1149.
11. Wei ZQ, Zhang YH, Ke CZ, et al. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World J Gastroenterol. 2017;23(34):6252–6260. doi:10.3748/wjg.v23.i34.6252
12. Moghadamtousi SZ, Kadir HA, Hassandarvish P, Tajik H, Abubakar S, Zandi K. A review on antibacterial, antiviral, and antifungal activity of curcumin. Biomed Res Int. 2014;2014:186864.
13. Pecheur EI. Curcumin against hepatitis C virus infection: spicing up antiviral therapies with ‘nutraceuticals’? Gut. 2014;63(7):1035–1037. doi:10.1136/gutjnl-2013-305646
14. Deljoo S, Rabiee N, Rabiee M. Curcumin-hybrid nanoparticles in drug delivery system (review). Asian J Nanosci Mat. 2019;2(1):66–91.
15. Farjadian F, Moghoofei M, Mirkiani S, et al. Bacterial components as naturally inspired nano-carriers for drug/gene delivery and immunization: set the bugs to work? Biotechnol Adv. 2018;36(4):968–985. doi:10.1016/j.biotechadv.2018.02.016
16. Jayakumar R, Selvamurugan N, Nair SV, Tokura S, Tamura H. Preparative methods of phosphorylated chitin and chitosan–an overview. Int J Biol Macromol. 2008;43(3):221–225. doi:10.1016/j.ijbiomac.2008.07.004
17. Akhtar F, Rizvi MM, Kar SK. Oral delivery of curcumin bound to chitosan nanoparticles cured Plasmodium yoelii infected mice. Biotechnol Adv. 2012;30(1):310–320. doi:10.1016/j.biotechadv.2011.05.009
18. Sun L, Chen YN, Zhou YL, et al. Preparation of 5-fluorouracil-loaded chitosan nanoparticles and study of the sustained release in vitro and in vivo. Asian J Pharm Sci. 2017;12(5):418–423. doi:10.1016/j.ajps.2017.04.002
19. Loutfy SA, El-Din HMA, Elberry MH, Allam NG, Hasanin MTM, Abdellah AM. Synthesis, characterization and cytotoxic evaluation of chitosan nanoparticles: in vitro liver cancer model. Adv Nat Sci. 2016;7:035008.
20. Ramana LN, Sharma S, Sethuraman S, Ranga U, Krishnan UM. Evaluation of chitosan nanoformulations as potent anti-HIV therapeutic systems. Biochim Biophys Acta. 2014;1840(1):476–484. doi:10.1016/j.bbagen.2013.10.002
21. Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi:10.1021/jm051197e
22. Schrödinger LNY. 2: Prime, Version 3. New York: Schrödinger, LLC. 2013. [computer program].
23. Tsutsumi Y. L2-solutions for nonlinear schrödinger equations and nonlinear groups. Funkcialaj Ekvacioj. 1987;31:115–125.
24. Nayeem A, Sitkoff D, Krystek S. A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci. 2006;15(4):808–824. doi:10.1110/ps.051892906
25. Adedeji AO, Singh K, Sarafianos SG. Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Cell Mol Biol. 2012;58(1):114–121.
26. Ibrahim SAL, Farroh KY, Elberry MH, Mohamed E-CB, Faraag AHI, Inventors; Arab Republic of Egypt, Ministry of Scientific Research. A method for preparation of anti HCV compounds against genotype 4a. 1120–2018, STDF ID: 15108.
27. Iraolagoitia XLR, Martini MF. Ca2+ adsorption to lipid membranes and the effect of cholesterol in their composition. Colloids Surf B. 2010;76(1):215–220. doi:10.1016/j.colsurfb.2009.10.037
28. Emam AN, Loutfy SA, Mostafa AA, Awad H, Mohamed MB. Cyto-toxicity, biocompatibility and cellular response of carbon dots–plasmonic based nano-hybrids for bioimaging. RSC Adv. 2017;7(38):23502–23514. doi:10.1039/C7RA01423F
29. Zhu ZJ, Ghosh PS, Miranda OR, Vachet RW, Rotello VM. Multiplexed screening of cellular uptake of gold nanoparticles using laser desorption/ionization mass spectrometry. J Am Chem Soc. 2008;130(43):14139–14143. doi:10.1021/ja805392f
30. Nunez R. DNA measurement and cell cycle analysis by flow cytometry. Curr Issues Mol Biol. 2001;3(3):67–70.
31. Karaliotas GI, Mavridis K, Scorilas A, Babis GC. Quantitative analysis of the mRNA expression levels of BCL2 and BAX genes in human osteoarthritis and normal articular cartilage: an investigation into their differential expression. Mol Med Rep. 2015;12(3):4514–4521. doi:10.3892/mmr.2015.3939
32. Zekri AR, Bahnassy AA, Hafez MM, et al. Characterization of chronic HCV infection-induced apoptosis. Comp Hepatol. 2011;10(1):4. doi:10.1186/1476-5926-10-4
33. Jiang XH, Xie YT, Cai YP, Ren J, Ma T. Effects of hepatitis C virus core protein and nonstructural protein 4B on the Wnt/β-catenin pathway. BMC Microbiol. 2017;17(1):124. doi:10.1186/s12866-017-1032-4
34. Jahromi MAM, Al-Musawi S, Pirestani M, et al. Curcumin-loaded chitosan tripolyphosphate nanoparticles as a safe, natural and effective antibiotic inhibits the infection of staphylococcus aureus and Pseudomonas aeruginosa in vivo. Iran J Biotechnol. 2014;12(3):1–8. doi:10.15171/ijb.1012
35. Das RK, Kasoju N, Bora U. Encapsulation of curcumin in alginate-chitosan-pluronic composite nanoparticles for delivery to cancer cells. Nanomedicine. 2010;6(1):153–160. doi:10.1016/j.nano.2009.05.009
36. Mandrol PS, Bhat K, Prabhakar AR. An in vitro evaluation of cytotoxicity of curcumin against human dental pulp fibroblasts. J Indian Soc Pedod Prev Dent. 2016;34(3):269–272. doi:10.4103/0970-4388.186757
37. Samrot AV, Burman U, Philip SA, Shobana N, Chandrasekaran K. Synthesis of curcumin loaded polymeric nanoparticles from crab shell derived chitosan for drug delivery. Inf Med Unlocked. 2018;10:159–182. doi:10.1016/j.imu.2017.12.010
38. Zhou N, Zan X, Wang Z, et al. Galactosylated chitosan-polycaprolactone nanoparticles for hepatocyte-targeted delivery of curcumin. Carbohydr Polym. 2013;94(1):420–429. doi:10.1016/j.carbpol.2013.01.014
39. Mirakabad FST, Akbarzadeh A, Milani M, et al. A comparison between the cytotoxic effects of pure curcumin and curcumin-loaded PLGA-PEG nanoparticles on the MCF-7 human breast cancer cell line. Artif Cell Nanomed B. 2016;44(1):423–430. doi:10.3109/21691401.2014.955108
40. Nair RS, Morris A, Billa N, Leong CO. An evaluation of curcumin-encapsulated chitosan nanoparticles for transdermal delivery. AAPS Pharm Sci Tech. 2019;20(2):69. doi:10.1208/s12249-018-1279-6
41. Chou TC, Talalay P. Generalized equations for the analysis of inhibitions of michaelis-menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur J Biochem. 1981;115(1):207–216. doi:10.1111/j.1432-1033.1981.tb06218.x
42. Kumar NP, Sharma P, Kumari SS, et al. Synthesis of substituted phenanthrene-9-benzimidazole conjugates: cytotoxicity evaluation and apoptosis inducing studies. Eur J Med Chem. 2017;140:128–140. doi:10.1016/j.ejmech.2017.09.006
43. Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging-US. 2016;8(4):603–619. doi:10.18632/aging.v8i4
44. Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3(3):e00149. doi:10.1002/prp2.149
45. Gesing A, Masternak MM, Wang F, Lewinski A, Karbownik-Lewinska M, Bartke A. Decreased expression level of apoptosis-related genes and/or proteins in skeletal muscles, but not in hearts, of growth hormone receptor knockout mice. Exp Biol Med (Maywood). 2011;236(2):156–168. doi:10.1258/ebm.2010.010202
46. Kusuma A, Colpitts CC, Schang LM, et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. J Hepatol. 2013;58:S473–S473. doi:10.1016/S0168-8278(13)61163-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.