Back to Journals » OncoTargets and Therapy » Volume 13
FOXN4 Inhibits Breast Cancer Progression By Direct Activation Of P53
Received 26 February 2019
Accepted for publication 21 July 2019
Published 7 January 2020 Volume 2020:13 Pages 71—81
DOI https://doi.org/10.2147/OTT.S206775
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
This paper has been retracted.
Hui Ye, 1 Meiling Duan 2
1Department of Galactophore, Linyi Central Hospital of Shandong, Linyi, People’s Republic of China; 2Department of Respiratory One, Linyi Central Hospital of Shandong, Linyi, People’s Republic of China
Correspondence: Meiling Duan
Department of Respiratory One, Linyi Central Hospital, 17th Jiankang Road, Yishui, Linyi City, Shandong 276400, People’s Republic of China
Email [email protected]
Background: Fork head domain-containing gene family (Fox) transcription factors, consisting of over 20 members, are involved in the progression of certain types of tumor. However, whether FOXN4 is involved in carcinogenesis and tumor progression is still unclear.
Purpose: In this study, we investigated the clinicopathological significance and the underlying mechanism of FOXN4 in breast cancer.
Methods and results: We examined the lower expression of FOXN4 in breast cancer tissues and cancer cell lines. The expression of FOXN4 is negatively correlated with tumor size and lymph node metastasis. Using CCK-8 assay, colony formation assay, wound healing assay, and Transwell assay, we revealed that FOXN4 notably decreased breast cancer cell proliferation, epithelial-mesenchymal transition and invasion in vitro. In addition, quantitative chromatin immunoprecipitation and luciferase assays determined that FOXN4 was able to directly bind with the promoter of P53. RT-qPCR and Western blotting analysis showed that FOXN4 could directly activate P53 expression. Functionally, P53 knockdown rescued the tumor inhibition effects of FOXN4 in breast cancer cells.
Conclusion: The present study provides new insights into the role of FOXN4 in breast cancer progression and suggests FOXN4 might represent a potential therapeutic target in breast cancer by modulating P53.
Keywords: FOXN4, breast cancer, proliferation, invasion, P53, transcription activation
Introduction
Breast cancer (BC) is a most common contributor leading to cancer death in females worldwide.1,2 The high mortality of breast cancer result from metastatic disease.3,4 Although significantly diagnosis and clinical treatment strategies have improved in the past decades, many patients with breast cancer still exhibit low survival and high recurrence.5,6 The precise potential molecular mechanism of breast cancer remains ambiguous and there is an urgent need to develop new therapeutic strategy.
The fork head domain-containing superfamily proteins (Fox) contains over 20 members in mammals and have important roles in different biological processes.7 The disturbance of fox proteins could result in the development of multiple kinds of disease.
As a subclass of Fox family transcription factors, FOXN has six members: FOXN1, FOXN2, FOXN3, FOXN4, FOXN5 and FOXN6.8,9 Among them, FOXN4 is the least known one.10 Although as reported that FOXN4 is required to activate Ptf1a, Neurod1, and Neurod4 expression for amacrine and horizontal cell generation,11 the function of FOXN4 remains largely unknown. In this study, we investigated the function of FOXN4, a potential tumor suppressor, through activation of P53, to inhibit tumor growth and invasion in breast cancer.
Materials And Methods
Tissue Samples Collection
80 paired breast cancer tissues, including their corresponding adjacent non-tumor tissues were collected from patients underwent tumor surgical resections in Linyi central hospital from January 2013 to July 2015. Patients received chemotherapy or radiotherapy before surgery were excluded. The tissue samples were fixed in 4% formalin at −80°C and embedded in paraffin until total RNAs were extracted. Informed consent and written approval were obtained from each patient. The study was approved by the Ethics Committee for Clinical Research of Linyi central hospital (Linyi, China).
Cell Culture And Transfection
MCF-7, MDA-MB-231, ZR-75-1, MDA-MB-453 and T47D cells were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM or RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37◦C and 5%CO2 in humidified air. The normal breast cell line MCF-10A was purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China), The MCF-10A cells were cultured in DMEM/F12, with 10% FBS, 20 ng/mL EGF, 0.1 mg/mL CT, 10 mg/mL insulin, and 500 ng/mL hydrocortisone (Sigma). The transfection was performed using the lipofectamine 3000 (Invitrogen, Carlsbad, CA), followed by the protocol recommended by the manufacturer. The plasmids used were synthesized by GenePharma (Shanghai, China). Knockdown of FOXN4 and P53 were achieved by using lentivirus carrying the relative shRNA sequences (Sigma). The establish confirmation of stable lentiviral transduction was determined using infection with shSCR as negative controls.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from breast cancer tissues and cells using TRIzol (Thermo Fisher Scientific, Inc.). 1 µg of total RNA in each group was reverse-transcribed in a volume of 20 µl using random and oligo dT primers, using the PrimeScript RT kit (Takara Biotechnology Co., Ltd., Dalian, China). RT-qPCR was conducted using SYBR-Green (Roche) mixture on an Applied Biosystems 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) Quantitative gene amplification was performed using the following thermocycling conditions: 40 cycles of pre-denaturation at 90°C for 30 sec, annealing at 90°C for 5 sec and final extension at 60°C for 30 sec. The 2−ΔΔCq method was employed for quantification. GAPDH was used as a reference gene. The primers used were as follows: E-cadherin forward 5-GAAATCACATCCTACACTGCCC-3, reverse 5-GTAGCAACTGGAGAACCATTGTC-3; Vimentin forward 5- ATTGAGATTGCCACCTACAG, reverse 5-ATCCAGATTAGTTTCCCTCAG-3; GAPDH forward 5-AGCCACATCGCTCAGACAC-3, reverse 5-GCCCAATACGACCAAATCC-3.
Western Blot Analysis
For Western blot, the cells were harvested and lysed using the radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China). The protein concentration was determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.). A total of 30 μg protein in each group was fractionated by SDS-PAGE (10% gel) and followed by transferred onto polyvinylidene fluoride membranes. After blocking with 5% fat-free milk for 1 h at room temperature, the membranes were incubated with indicated primary antibodies at 4°C overnight. After extensive washing with PBST for 10 min three times, HRP–conjugated anti-rabbit antibody (1:5000; cat no. 7074; Cell Signaling Technology, Inc.) or anti-mouse (1:10,000; cat no. 7076; CST) was added to the membrane and incubated at room temperature for another hour. Enhanced chemiluminescence (ECL Plus kit; Thermo) was used to visualize the protein bands. The X-ray films were scanned and analyzed using ImageJ 1.47i software (National Institutes of Health, Bethesda, MD, USA).
MTT Assay
The viability of breast cancer cells was determined by using the MTT assay. In brief, 2000 MCF-7 or MDA-MB-231 cells were seeded into a 96-well plate, incubated for 24 h, and separately transfected with the relative construct for 48 h. On each time point, 10 μl MTT reagent was added to each well and incubated for another 4 h. And then the medium of the well was removed and 200 μl of dimethyl sulfoxide was added to stop the reaction. The absorbance (OD value) at a wavelength of 490 nm was detected using a microplate reader for calculating cell viability.
Colony Formation Assay
Cell proliferation was analyzed using the plate colony formation assay. A total of 1000 cells were added to a 6-well plate and incubated in a humid incubator at 37°C with 5% CO2 and cultured for 14 days until colony formation was observed. The colonies were fixed with 100% methanol for 30 min and stained with 0.2% crystal violet (Beyotime Institute of Biotechnology, Haimen, China) for 15 min at room temperature. The colonies with more than 50 cells were counted and images were obtained under a microscope (magnification, x40, Olympus Corporation, Tokyo, Japan).
Migration Assays
Wound healing assay was used for migration analysis. The breast cancer cells transfected with the relative construct were cultured in DMEM medium containing with 10% FBS and maintained at 37 °C until 90% confluent. Linear scratches were created using a 20 ul pipette tip, and then the medium was replaced with serum-free DMEM medium for 24 h. The distance between two wounds was measured under an optical microscope and analyzed with Image J software (Bethesda, USA).
Invasion Assays
Invasion assays were analyzed by using Transwell chamber (8 μM pore size, Costar, Cambridge, MA, USA) according to the manufacturer’s instruction. The upper sides of the filters were coated with 100 μl Matrigel (BD Biosciences), 5×104 cells in each group were plated in the upper chamber with 600 µl of DMEM medium. While 1000 μl DMEM medium containing 20% FBS was added to the bottom chamber to act as the nutritional attraction. After incubation for 24 h, the cells on the upper membrane surface were removed, whereas transwell inserts were fixed in 4% paraformaldehyde for 15 min and then stained with 0.1% crystal violet for 10 min. Then, the cells were counted in 3 randomly selected fields of each chamber under a light microscope (Olympus Corporation). All experiments were repeated three times.
Dual Luciferase Reporter Assay
MCF-7 or MDA-MB-231 (3×104 cells/per well) cells were planted into 24-well plates for 24 h and co-transfected with the P53 WT or MUT promoter reporter plasmid, a Renilla luciferase plasmid and FOXN4 or siFOXN4 or NC. Firefly luciferase activity was measured 24 h after transfection using a dual luciferase assay kit, in accordance with the manufacturer’s protocol (Promega Corporation, Madison, WI, USA), and normalized to the corresponding Renilla luciferase activity.
Statistical Analysis
Statistical analysis was performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Each experiment was performed at least three times. Data was represented as means ± standard deviations (SD). Student’s t-test (two-tailed) was used to analyze the difference between two groups. One way ANOVA with post hoc test by Tukey’s test was used to analyze data of more than two groups. P<0.05 was considered to have a statistically significant difference.
Results
The Expression Of FOXN4 In Breast Cancer Tissues Is Low And Associated With Better Prognosis
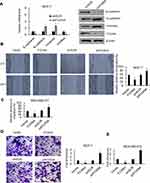
The expression profile of FOXN4 in breast cancer was not previously fully elucidated. In the present study, the expression pattern of FOXN4 in normal breast tissues and breast cancer tissue samples was examined using RT-qPCR. As shown, FOXN4 was significantly downregulated in the cancer sample compared with adjacent normal tissues (Figure 1A). We further detect the expression of FOXN4 in breast cancer using the online database of the human protein atlas. Similarly, the results illustrated that the protein expression of FOXN4 was predominantly lower in the cancer tissues (Figure 1B). Using the online database of The Cancer Genome Atlas (TCGA) with breast cancer, we identified the higher FOXN4 expression was association with better overall survival (Figure 1C). Consistently, the expression of FOXN4 mRNA was assessed in MCF-7, MDA-MB-231, ZR-75-1, MDA-MB-453, T47D and normal breast epithelial cells MCF-10A. As shown, FOXN4 mRNA in breast cancer cells was lower than that in MCF-10A cells (Figure 1D). Subsequently, we analyzed the correlation between FOXN4 expression and clinicopathological parameters. As shown in Table 1, the expression of FOXN4 was negatively associated with tumor size and lymph node metastasis (p<0.01). The above suggesting FOXN4 plays a potential suppression role in breast cancer.
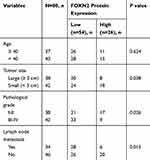 |
Table 1 Age And Clinicopathological Variables Of 80 BC Patients |
FOXN4 Inhibits Breast Cancer Cells Proliferation In Vitro
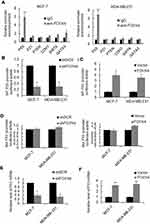
In order to further characterize the function of FOXN4 in breast cancer, we then constructed a lentivirus vector carrying the complete ORF of FOXN4 and established stable MCF-7-FOXN4 and MDA-MB-23-FOXN4 cell lines. The infection efficiency was verified using RT-qPCR assays with empty vector as controls (Figure 2A). Furthermore, we knockdown FOXN4 using shRNA in the two cell lines, and the transfection efficiency was determined using RT-qPCR (Figure 2B). To investigate the effect of FOXN4 on cellular proliferation, MTT assays were performed, which indicated that overexpression of FOXN4 inhibits breast cancer cells proliferation in vitro, while knockdown of FOXN4 significantly facilitates breast cancer proliferation (Figure 2C and D). The similarly tendency were also shown from the colony formation assay (Figure 2E and F).
FOXN4 Inhibits EMT And The Invasion Capability Of Breast Cancer Cells In Vitro
The effects of FOXN4 on breast cancer cell EMT and invasion were further explored. As shown in Figure 3A, In MCF-7 cells, knockdown of FOXN4 could result in the loss of Epithelial marker E-cadherin and the acquirement of mesenchymal marker such as N-cadherin and vimentin, which indicates FOXN4 could suppress EMT in breast cancer cells (Figure 3A). To evaluate the migration potential of MCF-7 and MDA-MB-231 cells, wound healing assay was performed, compared with the vector and NC groups, cell migration was inhibited in the FOXN4 infection group and promoted in the shFOXN4 transfection group (Figure 3B and C). Consistently, in MCF-7 and MDA-MB-231cells transfected with shFOXN4 or infected with FOXN4, trasnswell assay was performed in vitro. The overexpression of FOXN4 could significantly decrease the capable of invasion, while knockdown of FOXN4 obviously increased the number of invasive cells (Figure 3D and E). The above results suggested that FOXN4 reduced the EMT, migration and invasion of breast cancer cells in vitro.
FOXN4 Directly Binds To The P53 Promoter And Activates Its Activity
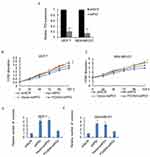
To elucidate the molecular mechanisms underlying the inhibition effect of FOXN4 in breast cancer progression, we identify potential key target genes in different pathway of FOXN4 in MCF-7 and MDA-MB-231 cells using qChIP assay, among them, the binding of P53 is the most remarkable one (Figure 4A). To verify that P53 is a direct target of FOXN4, luciferase reporter assays was carried out. The results illustrated that in MCF-7 and MDA-MB-231 cells transfected with shFOXN4, the luciferase activity was remarkedly reduced (Figure 4B). When FOXN4 was overexpressed, the relative P53 promoter luciferase activity was increased (Figure 4C). However, whether FOXN4 overexpression or knockdown had no effect on mutant type P53 promoter (Figure 4D). To further verify our hypothesis that P53 is a direct target for FOXN4, we examined the expression of P53 in MCF-7 and MDA-MB-231 cells transfected with FOXN4 shRNA or infected with FOXN4 overexpression construct. The data showed that inhibition of FOXN4 suppressed the mRNA level of P53, and ectopic expression of FOXN4 significantly improved the P53 expression (Figure 4E and F). The above finding suggests that FOXN4 directly activate P53 expression in breast cancer cells.
FOXN4 Inhibits The Proliferation Ability Of Breast Cancer Cells By Activating P53
Considering P53 as a target gene of FOXN4, we next investigated whether P53 knockdown could rescue the inhibition effect of FOXN4 in breast cancer cells. The knockdown efficiency of P53 was analyzed by RT-qPCR (Figure 5A). A rescue experiment was performed in MCF-7 and MDA-MB-231 cells. Results of MTT (Figure 5B and C) and colony formation assay (Figure 5D and E) showed that P53 knockdown could reverse the inhibition ability of FOXN4 in breast cancer cells viability and proliferation.
FOXN4 Suppresses The Migration And Invasion Of Breast Cancer By Regulating P53
To address whether the effects of FOXN4 in suppressing breast cancer cell migration and invasion can be attributed to its activation of P53. P53 was further knocked down in FOXN4 overexpression MCF-7 or MDA-MB-231 cells, a wound healing assay and transwell assay were performed respectively. Results of migration and invasion assay showed that P53 downregulation decreased the FOXN4 overexpression mediated migration MCF-7 cells (Figure 6A and B) and invasion of MCF-7 and MDA-MB-231 cells (Figure 6C and D). These results suggested that the effects of FOXN4 in the inhibition of breast cancer cells migration and invasion were activated by P53.
Discussion
A substantial body of evidence indicates that Fox family play important roles in a wide spectrum of biological processes as well as cancer progression.12–14 For example, FOXN1 is an attribute of benign epithelial tumors.15 FOXN4 is a tumor suppressor in breast cancer through inhibition of slug.16 Importantly, FOXN4 is a key regulator in a variety of biological processes during development. However, little is known about the function of FOXN4 in breast cancer.17,18 In this study, we showed that FOXN4 was downregulated in human breast cancer tissues and breast cancer cell lines compared with the adjacent normal tissues or the normal cell lines MCF-10A, suggesting the potential tumor suppressive role of FOXN4 in breast progression. The present study also demonstrated that knockdown of FOXN4 promoted breast cancer cell proliferation, EMT and invasion in MCF-7 and MDA-MB-231 cells.
In our previous study, we reported FOXO6 is downregulated in breast cancer, the function of which is to regulate migration, invasion, and epithelial-mesenchymal transition through transcriptionally regulated Sirt6 expression.19 As Li, W.20 reported that FOXN3 was a transcriptional repressor and promoted progression of hormonally responsive breast cancer through inhibition of GATA3. Interestingly, although we revealed that FOXN4 is another tumor driver, the mechanism is distinct. FOXN4 mainly function as transcriptional repressor to suppress P53 expression from mRNA and protein levels. P53, the most famous tumor suppressor, which controls several biological processes such as cell cycle, EMT, cell survival, metastasis and metabolism,21–23 is frequently inactivation in breast cancer and is considered a constant feature of cancer.24–26 In this study, we explored molecular changes associated with loss of p53 in breast cancer cell lines. Our results showed that FOXN4 could bind on the WT p53 promoter in MCF-7 and MDA-MB-231 cells and activate its expression. Interestingly, knockdown of FOXN4 could upregulate N-cadherin and vimentin and downregulate E-cadherin and thus promote EMT, which is a significant developmental process associated with cancer cell invasion.27 However, it is interesting to decipher the roles of FOXN4 in different types of breast cancers in the future depend on the expression of p53.
Consistent with previous studies, we provide direct evidence that different fox protein could be a distinct inducer or suppressor in the development of breast cancer. It would be interesting to determine if the FOXN4 mediated P53 regulation pathway can be a potential target for breast cancer therapy.
Availability Of Data And Materials
The analyzed data sets generated during the study are available from the corresponding author, on reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.21166
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi:10.1126/science.1203543
4. DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31–42. doi:10.3322/caac.21320
5. King TA, Morrow M. Surgical issues in patients with breast cancer receiving neoadjuvant chemotherapy. Nature Rev Clin Oncol. 2015;12(6):335–343. doi:10.1038/nrclinonc.2015.63
6. Vergine M, Scipioni P, Garritano S, et al. Breast-conserving surgery after neoadjuvant chemotherapy in patients with locally advanced cancer. Preliminary results. G Chir. 2013;34(9–10):254–256.
7. Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nature Rev Cancer. 2007;7(11):847–859. doi:10.1038/nrc2223
8. Schmidt J, Schuff M, Olsson L. A role for FoxN3 in the development of cranial cartilages and muscles in Xenopus laevis (Amphibia: Anura: Pipidae) with special emphasis on the novel rostral cartilages. J Anat. 2011;218(2):226–242. doi:10.1111/joa.2010.218.issue-2
9. Tribioli C, Robledo RF, Lufkin T. The murine fork head gene Foxn2 is expressed in craniofacial, limb, CNS and somitic tissues during embryogenesis. Mech Dev. 2002;118(1–2):161–163. doi:10.1016/S0925-4773(02)00220-4
10. Campbell EP, Quigley IK, Kintner C. Foxn4 promotes gene expression required for the formation of multiple motile cilia. Development. 2016;143(24):4654–4664. doi:10.1242/dev.143859
11. Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43(6):795–807. doi:10.1016/j.neuron.2004.08.041
12. Zhang W, Duan N, Song T, Li Z, Zhang C, Chen X. The Emerging Roles of Forkhead Box (FOX) proteins in osteosarcoma. J Cancer. 2017;8(9):1619–1628. doi:10.7150/jca.18778
13. Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nature Rev Genet. 2009;10(4):233–240. doi:10.1038/nrg2523
14. Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143(24):4558–4570. doi:10.1242/dev.112672
15. Grabowska AI, Wilanowski T. FOXN1 transcription factor in epithelial growth and wound healing. Mol Cell Biol. 2017;37:17. doi:10.1128/MCB.00110-17
16. Ye H, Duan M. FOXN2 is downregulated in breast cancer and regulates migration, invasion, and epithelial- mesenchymal transition through regulation of SLUG. Cancer Manag Res. 2019;11:525–535. doi:10.2147/CMAR.S176938
17. Boije H, Shirazi Fard S, Ring H, Hallbook F. Forkheadbox N4 (FoxN4) triggers context-dependent differentiation in the developing chick retina and neural tube. Differentiation. 2013;85(1–2):11–19. doi:10.1016/j.diff.2012.12.002
18. Boije H, Edqvist PH, Hallbook F. Temporal and spatial expression of transcription factors FoxN4, Ptf1a, Prox1, Isl1 and Lim1 mRNA in the developing chick retina. Gene Expression Patterns. 2008;8(2):117–123. doi:10.1016/j.modgep.2007.09.004
19. Ye H, Duan M. Downregulation of FOXO6 in breast cancer promotes epithelial-mesenchymal transition and facilitates migration and proliferation of cancer cells. Cancer Manag Res. 2018;10:5145–5156. doi:10.2147/CMAR.S157661
20. Li W, Zhang Z, Liu X, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. 2017;127(9):3421–3440. doi:10.1172/JCI94233
21. Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4(4):405–414. doi:10.1158/2159-8290.CD-13-0136
22. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–317. doi:10.1016/j.ccr.2014.01.021
23. Chang JW, Kuo WH, Lin CM, et al. Wild-type p53 upregulates an early onset breast cancer-associated gene GAS7 to suppress metastasis via GAS7-CYFIP1-mediated signaling pathway. Oncogene. 2018;37(30):4137–4150. doi:10.1038/s41388-018-0253-9
24. Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi:10.1073/pnas.0506230102
25. Shrestha M, Park PH. p53 signaling is involved in leptin-induced growth of hepatic and breast cancer cells. Korean J Physiol Pharmacol. 2016;20(5):487–498. doi:10.4196/kjpp.2016.20.5.487
26. Tsuchiya N, Izumiya M, Ogata-Kawata H, et al. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 2011;71(13):4628–4639. doi:10.1158/0008-5472.CAN-10-2475
27. Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi:10.1158/0008-5472.CAN-07-2017
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.