Back to Journals » OncoTargets and Therapy » Volume 13
Forkhead Box S1 Inhibits the Progression of Hepatocellular Carcinoma
Authors Lei D, Hu G, Chen Y, Hao T, Gao Y, Luo F
Received 22 July 2020
Accepted for publication 29 October 2020
Published 17 November 2020 Volume 2020:13 Pages 11839—11848
DOI https://doi.org/10.2147/OTT.S272596
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Dengliang Lei,1,2 Gangli Hu,1,2 Yue Chen,1,2 Tuantuan Hao,1,2 Yu Gao,1,2 Fang Luo1
1Department of Hepatobiliary Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Central Laboratory, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Fang Luo
Department of Hepatobiliary Surgery, The First Affiliated Hospital of Chongqing Medical University, 1 Youyi Road, Yuzhong, Chongqing 400016, People’s Republic of China
Email [email protected]
Introduction: Forkhead box (FOX) superfamily members were recently shown to play important roles in tumor development and progression. Forkhead box S1 (FOXS1), a member of the FOX family, has been reported to be closely associated with malignant neoplasms. However, its expression and effect on hepatocellular carcinoma remain unclear. The aim of this study was to determine the expression and role of FOXS1 in hepatocellular carcinoma (HCC).
Methods: Real-time PCR, Western blot and immunohistochemistry assays were carried out to determine FOXS1 expression in HCC tissues and cells. The biological roles of FOXS1 in HCC were investigated using CCK-8, colony formation, transwell and wound healing. Additionally, the effect of FOXS1 on epithelial–mesenchymal transition (EMT) was investigated by Western blotting. Xenograft model was carried out to evaluate the effect of FOXS1 in vivo.
Results: In our study, we confirmed lower FOXS1 expression in HCC samples than in normal liver tissues by performing Western blotting, immunohistochemistry and real-time PCR assays. In addition, FOXS1 expression is strongly associated with the prognosis of patients with HCC. Overexpression of FOXS1 suppressed cell proliferation, colony formation, the epithelial–mesenchymal transition (EMT) and the hedgehog (Hh) signaling pathway in vitro and in vivo. SAG, an activator of Hh signaling, partially reversed the effect of FOXS1 overexpression on HCC cells.
Conclusion: FOXS1 might suppress HCC cell proliferation, colony formation, and EMT by inhibiting the Hh signaling pathway, indicating that FOXS1 may be a promising biological target in HCC.
Keywords: FOXS1, epithelial–mesenchymal transition, hepatocellular carcinoma, hedgehog pathway
Introduction
Liver cancer is considered one of several cancers with the highest incidence rates worldwide,1 and HCC accounts for 80% of liver cancer cases.2 Despite advances in treatments such as surgery and ablation,2 liver cancer is still the fourth leading cause of death among all cancers,3 and thus, the molecular mechanisms of this lethal disease must be elucidated.
Forkhead box (FOX) proteins are an evolutionarily conserved family of transcriptional regulatory proteins that are closely related to the carcinogenesis and development of cancer.4 FOXS1, a member of this family, has been reported to be closely associated with stomach cancer and promotes cell proliferation and the epithelial–mesenchymal transition (EMT) in gastric cancer.5 Additionally, some researchers found that FOXS1 inhibits the proliferation, motility, and EMT of gastric cancer cells.6 FOXS1 is also associated with the differentiation of fibroblasts to myofibroblasts7 and the prognosis of patients with aggressive and metastatic breast cancer.8 Moreover, FOXS1 plays an important role in integrating and processing neuronal signals that are important for energy conversion and motor function.9 However, the mechanism underlying its effect on hepatocellular carcinoma has not been elucidated. In this study, we overexpressed FOXS1 in HCC cells to evaluate its function and found that FOXS1 suppressed the proliferation, motility and EMT of HCC cells by modulating the hedgehog (Hh) pathway.
Materials and Methods
Clinical Samples and Cell Lines
We collected 40 pairs of liver tumors and adjacent tissues from patients treated at The First Affiliated Hospital of Chongqing Medical University. Patients were excluded if they receive chemotherapy or radiation before surgery. Our study was approved by the Ethics Committee of Chongqing Medical University and that this was conducted in accordance with the Declaration of Helsinki. We obtained written informed consent from each patient after providing information about the study, and patients signed the informed consent forms. The normal hepatocyte cell line (HL-7702) and human HCC lines (SMMC-7721, Hep-3B, Hep-G2, SMMC-7402, and Huh-7) were purchased from the Institute of Chinese Academy of Sciences. SMMC-7721, Hep-3B, Hep-G2, SMMC-7402 and Huh-7 cell lines were cultured in DMEM (Corning Incorporated, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Thermo Scientific, Waltham, MA, USA). The HL-7702 cell line was cultured in RPMI 1640 medium (Corning Incorporated, Corning, NY, USA) supplemented with 10% FBS. All cells were cultured in an incubator with an atmosphere of 95% O2 and 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using TRIzol® reagent (Takara Biotechnology Co., Ltd., China) according to the manufacturer’s instructions. Reverse transcription and qRT-PCR were performed as previously described. The FOXS1 mRNA level was examined using real-time PCR. The sequences of the primers were as follows: FOXS1-forward, 5ʹ-AGCCCAAAGAGATTTCCACGC-3 and FOXS1-reverse, 5ʹ-CTGCTCCCGATGCCTGATTC-3ʹ.
Western Blot Analysis
Total proteins were extracted from HCC tissue samples or cells with RIPA lysis buffer (Beyotime Institute of Biotechnology, Jiangsu, China) containing protease inhibitors. Protein concentrations were measured with BCA protein detection kit (Beyotime Institute of Biotechnology, Jiangsu, China). Equal amounts of proteins were separated on 8% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with skimmed milk (dissolved in PBS) for 2 h, the membranes were subsequently probed using antibodies against β-actin (Cell Signaling Technology, USA, 1:10,000), FOXS1 (Sigma, 1:1000), vimentin (Cell Signaling Technology, USA, 1:1000), E-cadherin (Cell Signaling Technology, USA, 1:1000), N-cadherin (Cell Signaling Technology, USA, 1:1000), Slug (Cell Signaling Technology, USA, 1:1000), Hedgehog receptor protein patched homolog 1 (Cell Signaling Technology, USA, 1:1000), Sonic Hedgehog (Cell Signaling Technology, USA, 1:1000), smoothened (Cell Signaling Technology, USA, 1:1000) and glioma-associated oncogene homolog 1 (Cell Signaling Technology, USA, 1:1000) and glioma-associated oncogene homolog 1 (Cell Signaling Technology, USA, 1:1000) overnight at 4°C. The membranes were then washed with Tris-buffered saline containing Tween and incubated with an HRP-conjugated anti-rabbit antibody at 37°C for 2 h. Finally, the protein bands on the membranes were observed with an Odyssey Scanning System.
Immunohistochemistry (IHC)
We collected 40 human liver tumor tissues and adjacent tissues. The tissues were fixed with 4% paraformaldehyde, embedded in paraffin and sliced (3 μm). The samples were subjected to high-pressure antigen retrieval in pH 6.0 citrate buffer for 5 min, blocked with a buffer containing 10% bovine serum albumin for 60 min, and incubated with primary antibodies overnight at 4°C followed by an incubation with the peroxidase-conjugated anti-rabbit secondary antibody (1:500).
Transfection
Huh-7 and Hep-3B cells were infected with Lentivirus5-GFP-FOXS1. Transfected cells were cultured in complete media containing puromycin (1 μg/mL) for 72 h to generate a stable cell line overexpressing GFP-FOXS1. The methods were performed in accordance with the corresponding guidelines and regulations.
Cell Proliferation Assay
Huh-7 and Hep-3B cells were plated in 96-well plates at a density of 5×103/well. The Cell Counting Kit-8 (CCK8) cell proliferation assay was conducted according to the manufacturer’s protocol. Absorbance was measured at a wavelength of 450 nm.
Transwell and Wound Healing Assays
Transwell chambers were used to analyze the ability of cells to migrate and invade. A total of 5×104 cells in serum-free culture medium were directly and uniformly distributed in the well for the migration assay. Similarly, the invasion assay was performed almost identically to the migration assay, except that the upper chamber was first precoated with Matrigel according to the manufacturer’s instructions. For both assays, medium containing 15% FBS was added to the lower chamber as a chemoattractant. The cells were incubated in an atmosphere containing 5% CO2 for 12 (migration) or 16 h (invasion). The cells on the upper surface of the chamber were removed by wiping the membrane with a cotton swab, and invaded cells were fixed with 4% formaldehyde and stained with a 0.5% crystal violet solution. Cells were grown to confluence in 6-well plates. Six wounds were created in each well with sterile straw heads. The cells were washed with PBS and then cultured with DMEM. After 0 or 24 h of incubation, the same location in the wells was photographed to assess migration.
In vivo Xenograft Mouse Model
This experiment conforms to the the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health, and has been approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (approval number: 2020–645). Athymic nude mice (4 weeks old) were obtained from the Experimental Animal Center of Chongqing Medical University. Mice were subcutaneously injected with 5×106 HCC cells. Tumor size was monitored with calipers weekly. Mice were sacrificed 30 days after the injection, and subcutaneous tumor formation was observed.
Statistical Analysis
Data were analyzed using GraphPad Prism 7 software (GraphPad Software, Inc., San Diego, CA, USA) and SPSS 20.0 software and are reported as means ± standard errors of the means (SEMs). T-tests or analysis of variance were used for statistical comparisons. *P <0.05 was considered statistically significant.
Results
FOXS1 Was Expressed at Lower Levels in Tumor Tissues and Associated with a Poor Prognosis
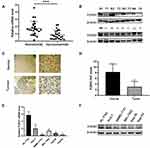
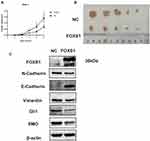
We conducted qRT-PCR, Western blotting and IHC to detect the levels of the FOXS1 mRNA and protein in paired samples of HCC and normal adjacent tissues.The FOXS1 mRNA was expressed at lower levels in HCC tissues than in the corresponding para-carcinoma tissues (Figure 1A). As shown in Figure 1B, the tumor tissue also expressed the FOXS1 protein at a significantly lower level. Immunohistochemical staining also revealed lower FOXS1 expression in HCC samples compared with adjacent normal liver samples (Figure 1C and D). Then, we detected FOXS1 expression in several human HCC cell lines. Compared with normal liver cells, the FOXS1 mRNA and protein were expressed at significantly lower levels in HCC cell lines (Figure 1E and F).
Finally, we collected 40 pairs of HCC tissues and their adjacent para-carcinoma tissues and assessed the clinicopathological features of patients to further examine the association between FOXS1 expression and clinicopathological factors. FOXS1 expression was related to the TNM stage (Table 1), suggesting that FOXS1 expression levels may be a biomarker for liver cancer.
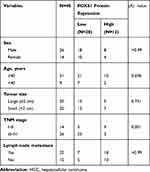 |
Table 1 Clinicopathological Characteristics of 40 Patients with HCC |
Overexpression of FOXS1 Suppresses the Proliferation and Colony Formation of HCC Cells
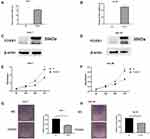
We overexpressed FOXS1 in two cell lines, Hep-3B and Huh-7, to elucidate the function of FOXS1 in HCC cells (Figure 2A–D). CCK8 and colony formation assays were performed to analyze cell proliferation. The cell growth curve showed a significant decrease in the proliferation of the experimental group compared with the control group (Figure 2E and F). In addition, overexpression of FOXS1 significantly reduced the number of colonies formed by the two cell lines (Figure 2G and H).
FOXS1 Inhibits the Migration and Invasion of HCC Cells
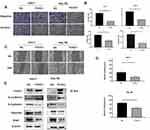
Transwell and wound healing experiments were conducted to investigate the effects of FOXS1 on the migration and invasion of HCC cells. FOXS1 overexpression inhibited the migration and invasion of Hep-3B and Huh-7 cell lines (Figure 3A and B). Similarly, in these two cell lines, the wound healing ability was lower than the control group when FOXS1 was overexpressed (Figure 3C and D). In addition, the EMT of Hep-3B and Huh-7 cells was inhibited by FOXS1 overexpression, as determined by the levels of E-cadherin, N-cadherin and other EMT-related proteins (Figure 3E).
FOXS1 Alters the EMT in HCC via the Hh Signaling Pathway
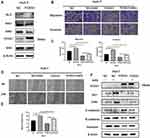
The Hh signaling pathway is closely related to tumor formation.10 Prior to this study, FOXS1 was reported to be associated with the Hh pathway.11 As shown in the figure, the levels of Sonic Hedgehog (Shh), Hedgehog receptor protein patched homolog 1 (PTCH1), smoothened (SMO) and glioma-associated oncogene homolog 1 (Gli1) were significantly decreased in the FOXS1-overexpressing Huh-7 cell line (Figure 4A). We used a SMO agonist to activate the Hh signaling pathway and further confirm whether FOXS1 modulates the EMT through the Hh signaling pathway. SAG induced SMO expression in FOXS1-overexpressing Huh-7 cells, and cell metastasis and invasion were significantly increased (Figure 4B–E). As shown in Figure 4F, SMO and GLI1 expression were significantly upregulated in the cell lines treated with the agonist, while E-cadherin levels were decreased and vimentin levels were increased. Thus, FOXS1 modulates the EMT through the Hh signaling pathway.
FOXS1 Inhibits Tumor Growth in vivo
A subcutaneous xenograft mouse model was used to assess the effect of FOXS1 on tumor growth in vivo. Huh-7 cells overexpressing FOXS1 and negative control cells were injected into the backs of nude mice. The tumor size was measured for 4 weeks (Figure 5A), and all the mice were sacrificed after 4 weeks. The tumor size in the experimental group were significantly smaller than in the control group (Figure 5B). Based on these results, FOXS1 inhibited tumor growth. In addition, the expression of SMO, GLI1, E-calcitonin and Vimentin was detected using Western blotting (Figure 5C).
Discussion
Despite the rapid development of treatments, the prognosis of patients with HCC remains poor, due to high recurrence and drug resistance rates.12 The metastasis of HCC also clearly decreases therapeutic efficacy in patients with HCC.13 Therefore, additional studies are urgently needed to obtain a better understanding of the mechanisms underlying the occurrence, development, and metastasis of HCC.
The liver plays an important role in the body. Hepatic transcription factors also play important roles in regulating the specific functions of the liver.4,5 Hepatic transcription factors include members of the FOX family, the alkaline leucine zipper family, the PP-homologous domain family (but not HNF1) and the nuclear orphan receptor family.14 Among these families, liver-enriched FOX transcription factors exert a significant effect on the development of HCC.15,16 FOXS1 plays a critical role in tumors.5,6,8 Wang reported FOXS1 overexpression in gastric cancer tissue through a mechanism mediated by GLI1 and miR-125a-5p, which promotes gastric cancer cell proliferation and the EMT.5 Interestingly, Lu discovered that FOXS1 expression is downregulated in GC, which promotes cell proliferation and metastasis by activating the Wnt/β-catenin pathway.6 Previous research reports that FOXS1/GLI1 interact in some tumors, and high FOXS1 expression is associated with a better prognosis.17 However, the definitive contribution of FOXS1 to the invasion and migration of HCC remains unclear. In the current study, we detected FOXS1 expression and observed a decrease in HCC tissues and cells. Then, we evaluated the effect of FOXS1 on the invasion and migration of HCC and subsequently explored the underlying mechanisms. FOXS1 inhibited the invasion and migration of Huh7 and Hep-3B cells, and FOXS1 also suppressed cell proliferation. Finally, FOXS1 overexpression inhibited the Hh signaling pathway, and when a SMO agonist was used to activate the Hh pathway, the effect of FOXS1 expression was partially reversed. Additionally, tumor growth was significantly reduced upon FOXS1 overexpression in nude mice, similar to the phenomena observed in vitro. Based on these results, FOXS1 may be a potential tumor suppressor.
The EMT is reported to play key roles in metastasis and invasion and may lead to the early metastasis of tumors.18 The EMT occurs during embryonic development to allow cells to transition between epithelial and mesenchymal states, and the transition to mesenchymal states modifies the expression of adhesion molecules in cells to promote their migration and metastasis.18 In addition, the EMT plays an important role in many biological processes, including wound healing, embryonic development, and fibrosis.19–21 Down-regulation of epithelial markers (E-cadherin) and up-regulation of mesenchymal markers (N-cadherin and vimentin) are the characteristics of the EMT,22 which play an important role in the early metastasis of cancer. The EMT plays an important role in HCC development.23 However, the relationship between FOXS1 and the EMT in HCC remains unclear. Here, overexpression of FOXS1 reduced E-cadherin expression, increased N-cadherin, vimentin and Slug expression, and inhibited cell invasion and migration in HCC.
The Hh signaling pathway is associated with the development and homeostasis of several organs.24,25 Hyperactivity associated with mutations of proteins in the Hh signaling pathway is also related to many malignancies.17 The Hh pathway has been shown to be associated with the initiation and maintenance of tumor growth, drug resistance and metastatic spread.17 For example, Hh signaling increases the proliferation and metastasis of human pancreatic cancer cells.26 Activation of the Hh pathway plays an important role in the development of prostate cancer and its progression.27 When FOXS1 was overexpressed, the expression of transcription factors related to Hh signaling were reduced. When an agonist of the Hh pathway was added to the treated cells, the invasion and metastasis were partially rescued, further confirming that FOXS1 modulated the EMT of HCC through the Hh pathway. FOXS1 may inhibit the expression of Gli1 by binding to the promoter region of Gli1, suggesting that Gli1 and FOXS1 may bind and inhibit each other’s expression, thus affecting the Hh pathway. In conclusion, FOXS1 participated in the EMT by modulating Hh signaling and inhibited HCC proliferation, indicating that FOXS1 plays a key role in HCC progression. Thus, FOXS1 might be a promising therapeutic target in HCC.
Highlights
- In this paper, FOXS1 was expressed at low levels in HCC and closely related to a poor prognosis of patients.
- FOXS1 overexpression inhibited HCC cell proliferation, migration, invasion and EMT.
- Mechanistically, the hedgehog (Hh) signaling pathway was involved in the FOXS1-mediated regulation of the invasion, migration and EMT of HCC.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81372481), the Science and Technology Research Program of Chongqing Municipal Education Commission (No. KJQN201800416), and the Basic and Advanced Research Project of Science and Technology Commission of Chongqing Municipality (No. cstc2018jcyjAX0162).
Disclosure
None of the authors have conflicts of interest to declare.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604.
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
4. Bach DH, Long NP, Luu TT, Anh NH, Kwon SW, Lee SK. The dominant role of forkhead box proteins in cancer. Int J Mol Sci. 2018;19(10):3279. doi:10.3390/ijms19103279
5. Wang S, Ran L, Zhang W, et al. FOXS1 is regulated by GLI1 and miR-125a-5p and promotes cell proliferation and EMT in gastric cancer. Sci Rep. 2019;9(1):5281. doi:10.1038/s41598-019-41717-w
6. Lu Q, Ma X, Li Y, et al. Overexpression of FOXS1 in gastric cancer cell lines inhibits proliferation, metastasis, and epithelial-mesenchymal transition of tumor through downregulating wnt/β-catenin pathway. J Cell Biochem. 2019;120(3):2897–2907. doi:10.1002/jcb.26821
7. Noizet M, Lagoutte E, Gratigny M, et al. Master regulators in primary skin fibroblast fate reprogramming in a human ex vivo model of chronic wounds. Wound Repair Regen. 2016;24(2):247–262. doi:10.1111/wrr.12392
8. Rokkam P, Gugalavath S, Gift Kumar DK, Vempati RK, Malla RR. Prognostic role of Hedgehog-GLI1 signaling pathway in aggressive and metastatic breast cancers. Curr Drug Metab. 2020;21(1):33–43. doi:10.2174/1389200221666200122120625
9. Montelius A, Marmigère F, Baudet C, Aquino JB, Enerbäck S, Ernfors P. Emergence of the sensory nervous system as defined by Foxs1 expression. Differentiation. 2007;75(5):404–417. doi:10.1111/j.1432-0436.2006.00154.x
10. Kalluri R, Zeisberg E. Controlling angiogenesis in heart valves. Nat Med. 2006;12(10):1118–1119. doi:10.1038/nm1006-1118
11. Diao Y, Rahman MF, Vyatkin Y, et al. Identification of novel GLI1 target genes and regulatory circuits in human cancer cells. Mol Oncol. 2018;12(10):1718–1734. doi:10.1002/1878-0261.12366
12. Vu NB, Nguyen TT, Tran LC, et al. Doxorubicin and 5-fluorouracil resistant hepatic cancer cells demonstrate stem-like properties. Cytotechnology. 2013;65(4):491–503. doi:10.1007/s10616-012-9511-9
13. Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216(5):1016–1026.
14. Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (Review). Oncol Rep. 2016;36(3):1199–1206. doi:10.3892/or.2016.4964
15. Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38(6):1331–1347. doi:10.1016/j.hep.2003.09.034
16. Kumar S, Batra A, Kanthaje S, Ghosh S, Chakraborti A. Crosstalk between microRNA-122 and FOX family genes in HepG2 cells. Exp Biol Med (Maywood). 2017;242(4):436–440. doi:10.1177/1535370216681548
17. Peer E, Tesanovic S, Aberger F. Next-generation Hedgehog/GLI pathway inhibitors for cancer therapy. Cancers (Basel). 2019;11(4).
18. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi:10.1016/j.cell.2009.11.007
19. Shaw TJ, Martin P. Wound repair: a showcase for cell plasticity and migration. Curr Opin Cell Biol. 2016;42:29–37. doi:10.1016/j.ceb.2016.04.001
20. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. doi:10.1016/j.cell.2016.06.028
21. Yang MC, Wang CJ, Liao PC, Yen CJ, Shan YS. Hepatic stellate cells secretes type I collagen to trigger epithelial mesenchymal transition of hepatoma cells. Am J Cancer Res. 2014;4(6):751–763.
22. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi:10.1016/j.jhep.2016.05.007
23. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi:10.1038/nrc2620
24. Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30(6):303–312. doi:10.1016/j.tips.2009.03.007
25. McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114.
26. Hao K, Tian XD, Qin CF, Xie XH, Yang YM. Hedgehog signaling pathway regulates human pancreatic cancer cell proliferation and metastasis. Oncol Rep. 2013;29(3):1124–1132. doi:10.3892/or.2012.2210
27. Sheng T, Li C, Zhang X, et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. doi:10.1186/1476-4598-3-29
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.