Back to Journals » Drug Design, Development and Therapy » Volume 15
Forkhead Box Protein O1: Functional Diversity and Post-Translational Modification, a New Therapeutic Target?
Authors Zhang X, Jiang L, Liu H
Received 2 February 2021
Accepted for publication 19 April 2021
Published 3 May 2021 Volume 2021:15 Pages 1851—1860
DOI https://doi.org/10.2147/DDDT.S305016
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Manfred Ogris
Xiaojun Zhang,1 Lusheng Jiang,2 Huimin Liu3
1Department of Cardiology, Shandong Rongjun General Hospital, Jinan, 250013, People’s Republic of China; 2Department of Emergency, Shandong Rongjun General Hospital, Jinan, 250013, People’s Republic of China; 3Blood Purification Center, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, 250011, People’s Republic of China
Correspondence: Huimin Liu
Blood Purification Center, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, No. 42, Wenhua West Road, Lixia District, Jinan, 250011, Shandong Province, People’s Republic of China
Tel +86 531 6861 7808
Email [email protected]
Abstract: Forkhead box protein O1 (FoXO1) is a transcription factor involved in the regulation of a wide variety of physiological process including glucose metabolism, lipogenesis, bone mass, apoptosis, and autophagy. FoXO1 dysfunction is involved in the pathophysiology of various diseases including metabolic diseases, atherosclerosis, and tumors. FoXO1 activity is regulated in response to different physiological or pathogenic conditions by changes in protein expression and post-translational modifications. Various modifications cooperate to regulate FoXO1 activity and FoXO1 target gene transcription. In this review, we summarize how different post-translational modifications regulate FoXO1 physiological function, which may provide new insights for drug design and development.
Keywords: forkhead box protein O1, post-translational modification, transcription
Introduction
The forkhead box (FoX) family mainly function as transcription factors characterized by a conserved DNA-binding domain (the Forkhead box).1 The human FoX O class (FoXOs) belong to FoX family and plays a crucial role in energy metabolism, differentiation, apoptosis, cellular proliferation, stress.2,3 FoXO subfamily includes four members: FoXO1, FoXO3, FoXO4, and FoXO6, which are encoded by four different genes and represent functional diversity in regulation of cellular processes.1,3 FoXO1 is a transcription factor involved in the regulation of a wide variety of physiological process including glucose metabolism, lipogenesis, apoptosis, and autophagy.4–7 FoXO1 dysfunction is involved in the pathophysiology of various diseases including metabolic diseases, atherosclerosis, and tumors.2,8–10 The Human FoXO1 gene is located at chromosomal location 13q14.11 and encodes a 655 amino acid (aa) protein.11–13 FoXO1 tissue expression is ubiquitous and within the cell FoXO1 distribution is cytoplasmic or nuclear. The cellular distribution of FoXO1 depends on the internal environment and homeostasis. Nuclear FoXO1 binds to cis-acting response elements of downstream target genes to mediate its transcription-regulatory function.12,14–16
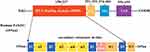
The FoXO1 protein comprises several functional domains, or motifs, including a conserved NH2-terminal DNA binding domain (DBD, residues 158–237), a nuclear localization signal motif (NLS, residues 251–253), a nuclear export sequence motif (NES, residues 374–401), and a COOH-terminal transcription activation domain (TAD, residues 596–655).1 DBD of FoXO1 is to recognize and bind to the consensus sequence of target gene promoter to activate transcription. NLS motif is responsible for transportation into the nucleus for transactivation, conversely, NES motif is to mediate transportation into the cytoplasm interacting with nuclear exportin-1 and 14-3-3 protein.17 The FoXO1 TAD can bind to other regulatory proteins to co-repress or coactivate its function.18 FoXO1 secondary structure includes four α-helices (α1, α2, α3, and α4), four β-strands (β1, β2, β3, and β4), one β turn, and two wing regions (W1 and W2). The domain or motif order in the protein is β1-α1-β2-α2-α4-α3-β3-β turn-wing 1-β4-wing 2, located at the N-terminus (residues 156–249) (Figure 1).1,15,18 Mutations or covalent alterations of residues may significantly affect FoXO1 conformation and flexibility, resulting in a functional change.19
FoXO1 function is regulated in response to different physiological or pathogenic conditions at transcription and post-translational levels.20–27 Modulators of FoXO1 activity have presented promising therapeutic value in the treatment of diabetes and obesity.28–31 In this brief review, to best understand the molecular regulatory mechanism modulating FoXO1 activity, we summarize how different post transcriptional modifications (PTMs) regulate FoXO1 physiological function. This review may provide new insights for drug design and development.
PTM of FoXO1 and the Effects on Protein Activity
PTM is an essential way to regulating protein function and control fundamental physiological processes. A range of modifications are involved in regulating FoXO1 activity, including phosphorylation, acetylation, methylation, ubiquitination, glcNAcylation, and glutathionylation. These modifications modulate FoXO1 activity by affecting its subcellular distribution, DNA binding affinity, or gradation.
Phosphorylation
Phosphorylation modification is the most common type of covalent PTM involved in regulating protein function. Phosphorylation can modulate protein localization, conformation, turnover, DNA binding, and protein-protein interactions.32 Phosphorylation site(s) are found within domains or motifs that are phosphorylated by specific protein kinases and can be associated with specific cellular processes.
Phosphoinositide 3-Kinase/Protein Kinase B (PI3K/PKB)
FoXO1 is a critical mediator of the insulin signaling pathway regulation of metabolic homeostasis in the liver and pancreatic β-cells in response to glucose alterations or stress.5,14,33 Phosphoinositide 3-kinase/protein kinase B (PI3K/PKB or Akt) is the main upstream kinase in FoXO1 signaling transduction pathway regulation. PKB is a negative regulator that phosphorylates FoXO1 at specific residues, including Threonine 24 (Thr24), Serine 256 (Ser256), and Serine 319 (Ser319).14,22,33–47 The Thr24/Ser256 residue is within the peripheral region of the DBD, adjacent to the NLS, indicating that phosphorylation may affect FoXO1 subcellular localization or NLS function.43,48 The FoXO1 Ser319 residue is between the NLS and NES and its phosphorylation potentially promotes nuclear export.40,48
FoXO1 is cytoplasmic in unstimulated β-cells, and translocates to the nucleus when exposed to H2O2 or hyperglycemia.33,49 In the liver, or other tissue, under basal conditions, FoXO1 is largely nuclear and transports into the cytoplasm with insulin treatment. However, FoXO1 insulin-stimulated translocation is inhibited by mutations in Thr24, Ser256, and Ser319.14,44,47,50 Insulin inhibits gluconeogenesis in the liver in a PKB-dependent manner. This leads to decreased expression of the key gluconeogenic enzymes, phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6P). Interestingly, nuclear FoXO1 can bind the insulin response element (IRE) in the PEPCK and G6P promoters.14,43
PKB-induced FoXO1 phosphorylation promotes cytoplasmic localization 14-3-3 protein binding, leading to reduced nuclear FoXO1 protein levels and FoXO1-mediated gene transcription in insulin/IGF-1 (insulin-like growth factor-1) signaling pathways.14,35,43,51,52 The 14-3-3 protein is a scaffold protein that sterically binds FoXO1. The FoXO1 binding region, residue 250–262, of FoXO1 is important for 14-3-3 protein binding and FoXO1 DNA binding ability decreases when FoXO1 is 14-3-3 bound, conformational change is possibly involved in the process.22,43,48,51,53 The PTM may result in protein conformational change and affect functional activity through influencing molecular weight, hydrogen bond, water solubility or flexibility.54 Saline et al report that the phosphorylated Ser22 and Thr24 unchanged the local secondary structure of the FoXO1, therefore, further study is needed to investigate the effect of phosphorylation at other residues on FoXO1 conformation.
Furthermore, in endothelial cells, dephosphorylation FoXO1 at Ser256 by peptidyl-prolyl isomerase (PPIase) leads to FoXO1 accumulation in the nucleus and increased transcription of genes involved in chemotaxis and apoptosis. These observations suggest that FoXO1 may play an important role in cardiovascular diseases.55 Additionally, insulin-PKB-induced FoXO1 phosphorylation and cytoplasmic accumulation are necessary for subsequent polyubiquitination and proteasome-mediated degradation.56
AMP-Activated Protein Kinase (AMPK)
AMP-activated protein kinase (AMPK) is a key regulator of energy homeostasis and is involved in regulating FoXO1 functions including in oxidative stress, glucose metabolism, tumorigenesis, and lifespan extension.57–61 AMPK can directly phosphorylate FoXO1 on residues Thr166, Ser202, Ser314, Ser321, Thr463, and Ser466. In C. elegans, AMPK-mediated FoXO1 phosphorylation enhances FoxO-dependent transcriptional activity and has beneficial effects of lifespan extension.61 The effect of individual phosphorylation sites requires additional investigation. In the hypoxic environment of pulmonary artery smooth muscle, AMPK is activated to increase FoXO1 nuclear localization and catalase expression.57 A study examining the regulation of hepatic gluconeogenesis reported that transforming growth factor β-induced activation of protein phosphatase 2A could dephosphorylate AMPK phosphorylated FoXO1 to promote FoXO1 nuclear translocation, but the phosphorylated residues involved were not addressed.60
Yun et al reported that AMPK phosphorylated the FoXO1 TAD domain located Thr649 residue to increase FoXO1 nuclear localization and transcriptional activity to combat oxidative stress in CHANG liver cells.58 Recent evidence shows that AMPK-mediated phosphorylation of Ser22 in the FoXO1 NH2-terminal domain reduces PKB-induced Thr24 phosphorylation. This results in inhibition of chaperone protein (14-3-3) binding, activating FoXO1-dependent transcriptional activity.22
MST1
The mammalian sterile 20-like kinase 1 (MST1) is involved in the regulation of cell death in response to oxidative stress. Early studies have demonstrated that MST1 phosphorylates the Ser207 of FoXO3, equivalent to FoXO1 Ser212 in the DBD, leading to FoXO3 nuclear accumulation.62 Consistent with this study, a more recent study reports that MST1 induces Ser212 FoXO1 phosphorylation and promotes FoXO1 nuclear accumulation through inhibiting the interaction with 14-3-3 proteins.63 MST1-induced phosphorylation of FoXO1 has a protective effect against ischemia or reperfusion in cardiomyocytes.64,65
In regulatory T cells (Treg), phosphorylation of FoXO1 Ser212 by MST1 facilitates the FoXO1 stability and promotes Treg cell development and function.66 Recent studies demonstrate that MST1 mediated endothelial angiogenesis occurring through FoXO1 Ser212 phosphorylation enhances FoXO1 nuclear localization.67 However, these findings were not consistent with those presented by Brent et al, who demonstrated that the phosphorylated serines could block FoXO1 binding to cis-acting elements of target genes.15
CDK1/2, CDK4
The cyclin-dependent kinase 1 (CDK1) phosphorylates FoXO1 at Ser249 in the W2 region and promotes nuclear translocation, leading to stimulation of FoXO1 transcription in postmitotic neurons without affecting its DNA binding ability.15,68 Nevertheless, both CDK1 and CDK2 can phosphorylate FoXO1 Ser249 and induce cytoplasmic localization resulting in inhibition of FoXO1 transcription in LNCaP and DU145 cells, showing a regulatory role in apoptosis and DNA damage.69,70 These differential effects indicate that CDK1/2 play different roles in various biological and pathogenetic processes. However, CDK4 phosphorylates FoXO1 Ser329 and inhibits its activity resulting in a decrease of FoXO1-induced recombination activating protein (Rag) expression to play a major role in B cell differentiation and genomic instability.71,72
Many other phosphorylation sites are also involved in the FoXO1 subcellular localization and transcriptional activity regulation. Protein kinase A (PKA) also phosphorylates FoXO1 at Ser276 (between NLS and NES), increasing its nuclear localization and stability and increasing gluconeogenesis.45 The casein kinase 1 (CK1) phosphorylates FoXO1 at the proximal Ser322 and Ser325 residues in the region between NLS and NES and promotes FoXO1 cytoplasmic localization and nuclear export through interaction with the chromosomal region maintenance protein-1 (CRM1).40 FoXO1 Ser329 is phosphorylated by the dual specificity tyrosine phosphorylated regulated kinase 1A (DYRK1A). This promotes cytoplasmic FoXO1 distribution in HEK-293 cells, which has an inhibitory effect on FoXO1 activity.73 Nemo-like Kinase (NLK) is kinase the functions upstream of FoXO1 and can phosphorylate mouse FoXO1 at Ser284, Ser295, Ser326, Ser380, Ser391, Thr399, Ser413, and Ser415 in TIGK cells. NLK phosphorylates human FoXO1 at Ser329 and decreases FoXO1 nuclear localization. NLK-mediated FoXO1 phosphorylation also inhibits the FoXO1-mediated expression of zinc finger E-box-binding homeobox 2 (ZEB2), a transcriptional inhibitor that regulates nervous system development and inflammatory responses.74–76
Mitogen-activated protein kinase (MAPK) is also involved in controlling FoXO1 activity. Extracellular signal-regulated protein kinase (Erk) and p38 MAPK (p38) directly phosphorylate FoXO1 on Ser246, Ser284, Ser295, Ser326, Ser413, Ser415, Ser429, Ser467 Ser475, and Ser284, Ser295, Ser326, Ser467, Ser475, respectively, and regulate FoXO1-induced angiogenic genes.77
Overall, there are numerous studies describing the phosphorylation of FoXO1 residues by upstream kinases and the corresponding effects on FoXO1 function. However, the physiological implication of other phosphorylated residues remains to be determined.76,77
Acetylation
The transcriptional regulation of FoXO1 is also involves modification by protein acetylation. The cAMP response element-binding protein-binding protein (CBP) and its homolog, p300 (CBP/p300) are histone acetyl transferases involved in the regulation of multiple pathophysiological processes such as oxidative stress.78,79 CBP binds and acetylates FoXO1 at lysine245 (Lys245), Lys248, and Lys262, near to the DBD in the wing 2 region (residues 244–249), and negatively regulates FoXO1 transcriptional activity in mammalian cells.80 These three acetylated lysines, as well as Lys265 acetylated by CBP/p300, induce charge changes in the protein and decrease the affinity of FoXO1 binding to target gene cis-acting elements, including G6P.15,81 Inconsistent with prior studies, Perrot et al shows that p300 directly acetylates FoXO1 and enhances the nuclear localization and transactivation of FoXO1 under basal conditions, and that this effect is reversed by insulin stimulation.82 Similar results have shown that p300 acetylates FoXO3a and enhances its transcriptional activity.83 The specific p300 acetylation site(s) were not identified in the study, and the divergent effects of CBP/p300 on FoXO1 function need further exploration.82
FoXO1 acetylation at Lys245, Lys248, Lys262, Lys265, and Lys274 upregulates c-Myc expression and promotes apoptosis in GBM cells. This effect is suppressed by Class IIa histone deacetylase (HDAC), and is not dependent on Akt-induced phosphorylation.84 By contrast, HDAC9, a HDAC subtype, deacetylates FoXO1 and enhances its transcriptional activity by upregulating gluconeogenic enzymes in the liver.24
Single PTMs can synergistically combine with other PTMs to coactivate, co-repress, or antagonize each other. Acetylated FoXO1 can facilitate FoXO1 phosphorylation through PI3K/PKB by the cascade amplification effect. This leads to increased cytoplasmic translocation, which indirectly promotes FoXO1 degradation and represents an overlying inhibitory effect on FoXO1 activity.81,85 Conversely, acetylated FoXO1 can be deacetylated by silent information regulator 1 (SirT1) and SirT2, which promote its nuclear localization and enhance transcriptional activity.23,80,86–89 SirT1 deacetylates FoXO1 and increases its activity and degradation through the ubiquitin-proteasomal degradation pathway in βTC-3 cells under hyperglycemia or oxidative stress conditions.49
When cytosolic FoXO1 is acetylated at Lys262, Lys265, and Lys274 it disassociates from SirT2 and interacts with an E1-like protein (atg7) to modulate autophagic process. These may have implications for human colon cancer therapies.4 FoXO1 CoRepressor (FCoR) is a novel FoXO1-binding protein expressed in mouse adipocytes. FCoR directly acetylates mouse FoXO1 Lys259, Lys 262, Lys 271, and Lys 291 (Lys262, Lys 265, Lys 274, and Lys 294 of human FoXO1) in vitro through disrupting the FoXO1 and Sirt1 interaction, resulting in decreased FoXO1-dependent gene expression and adiposity.90 The beneficial effect of FCoR on insulin sensitivity and adiposity is helpful for exploring therapeutic targets for the prevention of human obesity and type 2 diabetes.
Accumulating evidence demonstrate that acetylation participates in the regulation of FoXO1 function. Further research could explore how different acetyltransferases function on the same lysine residues, how the different modifications interact, and the functional roles of some acetylated lysines, such as the Lys597 in the FoXO1 TAD, newly identified through mass spectrometry.
Methylation
Protein arginine methyltransferases 1 (PRMT1) methylates the arginyl residue guanidino nitrogens of multiple proteins involved in the regulation of cellular processes including glucose metabolism and stress tolerance.91–93 In mice, FoXO1 is methylated by PRMT1 at Arg248 (Arg248) and Arg250 (equivalent to Arg251 and Arg253 in the human FoXO1 NLS). These residues are near Ser253 (Ser256 in human), and their methylation has an inhibitory effect on the Akt-mediated phosphorylation of FoXO1 Ser253. This leads to increased FoXO1 nuclear retention and transcriptional activity in response to oxidative stress-induced apoptosis.91,93,94
A recent study demonstrated that the Lys273 residue of FoXO1 is also methylated by euchromatic histone lysine methyltransferase 2 (EHMT2), which is a histone methyltransferase to regulate apoptotic process and cell differentiation.27,95 This methylation decreases FoXO1 stability through S-phase kinase-associated protein 2 (SKP2) mediated- ubiquitination degradation in colon cancer.27 The differential effect of methylation of different FoXO1 residues on protein function suggests biological regulatory complexity.
Ubiquitination
FoXO1 activity is controlled by the ubiquitination process. FoXO1 is degraded via the 26S ubiquitin-proteasome pathway in HepG2 and INS-1 cells, and polyubiquitination-mediated degradation is promoted by insulin through PI3K/PKB-induced phosphorylation, which leads to cytoplasmic translocation.56,96 SKP2, an E3 ubiquitin ligase, is involved in ubiquitination-mediated degradation and inhibition of FoXO1 activity through interacting with PKB-induced phosphorylation at Ser256.97 The E3 ubiquitin-protein ligase Mdm2 is also involved in FoXO1 ubiquitination-mediated degradation requiring the PKB-mediated phosphorylation.98
Intriguingly, ubiquitin-specific protease 7 (USP7) directly deubiquitinates monoubiquitinated FoXO1 and has an inhibitory effect on FoXO1 transcriptional activity, resulting in decreased expression of downstream genes including G6Pase and PEPCK.99 USP7 does not affect FoXO1 protein levels and subcellular localization, but decreases FoXO1 DNA binding affinity.99
GlcNAcylation
GlcNAcylation as a Ser/Thr residue-targeted post-translational protein modification involved in the regulation of glucose metabolism, oxidative stress, and tumorigenesis.100–102 FoXO1 GlcNAcylation by O-GlcNAc transferase (OGT) increases FoXO1-induced G6Pase expression, and this effect is not associated with FoXO1 protein expression, PKB-mediated phosphorylation, or subcellular distribution.103 Hyperglycemia increases FoXO1 GlcNAcylation and FoXO1 transcriptional activity, suggesting that FoXO1 GlcNAcylation plays an important role in diabetic pathophysiology.103–106 Reports indicate that the Thr317, Ser318, Ser550, Thr648, and Ser654 residues are the FoXO1 GlcNAcylated sites and that GlcNAcylated Thr317 affects the transcriptional activity of human FoXO1.104 However, Fardini et al revealed that this effect is not obvious in mouse FoXO1.107
Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1 α) is not only a GlcNAcylated protein but also a coactivator that interacts with OGT to enhance FoXO1 GlcNAcylation and subsequent transcriptional activity.108 Further work is needed to investigate the molecular mechanisms involved in GlcNAcylation and the regulatory functions through which GlcNAcylation may present as potential therapeutic target for drug development.109
Glutathionylation
Cysteine S-glutathionylation is post-translational protein modification that plays an important role in the physiological reaction to oxidative stress.110,111 FoXO1 glutathionylation mainly enhances FoXO1 DNA-binding capacity and subsequent transcriptional activity in response to oxidative environments, and does not affect FoXO1 phosphorylation status and subcellular location.112 However, which of the specific cysteine residues contribute to the effect and how remain unknown.
Perspectives
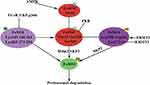
FoXO1 as a key transcription factor that participates in physiological processes including glucose metabolism, apoptosis, adipocyte differentiation, and oxidative stress. FoXO1 dysfunction contributes to a wide variety of physiopathological processes including tumor progression, Type 2 diabetes mellitus, obesity, and atherosclerosis.5,7,9,33,113–116 FoXO1 activity is closely linked with protein expression and is regulated by PTMs, including phosphorylation, acetylation, ubiquitination, methylation, glcNAcylation, and glutathionylation (Table 1). A growing body of evidence indicates that various modifications interact with each other or other coactivators (corepressors) to regulate FoXO1 activity and the transcription of FoXO1-mediated target genes (Figure 2).81,97,108 However, the regulatory role of modified FoXO1 in various diseases and cellular processes remain incompletely understood. Considerably more work will need to be done to fully understand the underlying molecular mechanisms of FoXO1 function, and the results of such studies will provide further insight into drug development, such as FCoR agonists, for human diseases.90,117–119
 |
Table 1 Human FoXO1 Posttranslational Modifications |
Disclosure
The authors report no conflicts of interest in this work.
References
1. Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27(16):2263–2275. doi:10.1038/onc.2008.20
2. Wang Y, Zhou Y, Graves DT. FOXO transcription factors: their clinical significance and regulation. Biomed Res Int. 2014;2014:1–13. doi:10.1155/2014/408514
3. Link W. Introduction to FOXO biology. FOXO Transcript Factors. 2019;1890:1–9. doi:10.1007/978-1-4939-8900-3_1
4. Zhao Y, Yang J, Liao W, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–675. doi:10.1038/ncb2069
5. Peng S, Li W, Hou N, Huang N. A review of FoxO1-regulated metabolic diseases and related drug discoveries. Cells. 2020;9(1). doi:10.3390/cells9010184
6. Crunkhorn S. Diabetes: selective FOXO1 modulation. Nat Rev Drug Discov. 2017;16(12):828.
7. Lu H, Huang H. FOXO1: a potential target for human diseases. Curr Drug Targets. 2011;12(9):1235–1244. doi:10.2174/138945011796150280
8. Cheng Z, Guo S, Copps K, et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15(11):1307–1311. doi:10.1038/nm.2049
9. Valenti L, Rametta R, Dongiovanni P, et al. Increased expression and activity of the transcription factor FOXO1 in nonalcoholic steatohepatitis. Diabetes. 2008;57(5):1355–1362. doi:10.2337/db07-0714
10. Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi:10.1038/sj.onc.1209086
11. Dunham A, Matthews LH, Burton J, et al. The DNA sequence and analysis of human chromosome 13. Nature. 2004;428(6982):522–528. doi:10.1038/nature02379
12. Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47(2):187–199. doi:10.1006/geno.1997.5122
13. Galili N, Davis RJ, Fredericks WJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5(3):230–235. doi:10.1038/ng1193-230
14. Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108(9):1359–1367. doi:10.1172/JCI200112876
15. Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008;16(9):1407–1416. doi:10.1016/j.str.2008.06.013
16. Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274(24):17184–17192. doi:10.1074/jbc.274.24.17184
17. Brunet A, Kanai F, Stehn J, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156(5):817–828. doi:10.1083/jcb.200112059
18. Zhao HH, Herrera RE, Coronado-Heinsohn E, et al. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem. 2001;276(30):27907–27912. doi:10.1074/jbc.M104278200
19. Psenakova K, Kohoutova K, Obsilova V, Ausserlechner M, Veverka V, Obsil T. Forkhead domains of FOXO transcription factors differ in both overall conformation and dynamics. Cells. 2019;8(9):966. doi:10.3390/cells8090966
20. Banks AS, Kim-Muller JY, Mastracci TL, et al. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 2011;14(5):587–597. doi:10.1016/j.cmet.2011.09.012
21. Goitre L, Balzac F, Degani S, et al. KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS One. 2010;5(7):e11786. doi:10.1371/journal.pone.0011786
22. Saline M, Badertscher L, Wolter M, et al. AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins. J Biol Chem. 2019;294(35):13106–13116. doi:10.1074/jbc.RA119.008649
23. Sin TK, Yung BY, Siu PM. Modulation of SIRT1-Foxo1 signaling axis by resveratrol: implications in skeletal muscle aging and insulin resistance. Cell Physiol Biochem. 2015;35(2):541–552. doi:10.1159/000369718
24. Chen J, Zhang Z, Wang N, et al. Role of HDAC9-FoxO1 axis in the transcriptional program associated with hepatic gluconeogenesis. Sci Rep. 2017;7(1):6102. doi:10.1038/s41598-017-06328-3
25. Nakae J, Kitamura T, Kitamura Y, Biggs WH, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4(1):119–129. doi:10.1016/S1534-5807(02)00401-X
26. Kappel BA, Stohr R, De Angelis L, Mavilio M, Menghini R, Federici M. Posttranslational modulation of FoxO1 contributes to cardiac remodeling in post-ischemic heart failure. Atherosclerosis. 2016;249:148–156. doi:10.1016/j.atherosclerosis.2016.04.001
27. Chae Y-C, Kim J-Y, Park JW, et al. FOXO1 degradation via G9a-mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019;47(4):1692–1705. doi:10.1093/nar/gky1230
28. Peng S, Xiao W, Ju D, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. 2019;11:488. doi:10.1126/scitranslmed.aau7116
29. Nagashima T, Shigematsu N, Maruki R, et al. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78(5):961–970. doi:10.1124/mol.110.065714
30. Zou P, Liu L, Zheng L, et al. Targeting FoxO1 with AS1842856 suppresses adipogenesis. Cell Cycle. 2014;13(23):3759–3767. doi:10.4161/15384101.2014.965977
31. Tanaka H, Nagashima T, Shimaya A, Urano Y, Shimokawa T, Shibasaki M. Effects of the novel Foxo1 inhibitor AS1708727 on plasma glucose and triglyceride levels in diabetic db/db mice. Eur J Pharmacol. 2010;645(1–3):185–191. doi:10.1016/j.ejphar.2010.07.018
32. Humphrey SJ, James DE, Mann M. Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol Metabol. 2015;26(12):676–687. doi:10.1016/j.tem.2015.09.013
33. Martinez SC, Cras-Meneur C, Bernal-Mizrachi E, Permutt MA. Glucose regulates Foxo1 through insulin receptor signaling in the pancreatic islet beta-cell. Diabetes. 2006;55(6):1581–1591. doi:10.2337/db05-0678
34. Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274(24):17179–17183. doi:10.1074/jbc.274.24.17179
35. Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi:10.1016/S0092-8674(00)80595-4
36. Del Peso L, Gonzalez VM, Hernandez R, Barr FG, Nunez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18(51):7328–7333. doi:10.1038/sj.onc.1203159
37. Biggs WH, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96(13):7421–7426. doi:10.1073/pnas.96.13.7421
38. Zhang X, Gan L, Pan H, et al. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277(47):45276–45284. doi:10.1074/jbc.M208063200
39. Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19(5):989–996. doi:10.1093/emboj/19.5.989
40. Rena G, Woods YL, Prescott AR, et al. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21(9):2263–2271. doi:10.1093/emboj/21.9.2263
41. Tang ED, Nunez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274(24):16741–16746. doi:10.1074/jbc.274.24.16741
42. Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52(3):642–649. doi:10.2337/diabetes.52.3.642
43. Schmoll D, Walker KS, Alessi DR, et al. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275(46):36324–36333. doi:10.1074/jbc.M003616200
44. Nakae J, Kitamura T, Ogawa W, Kasuga M, Accili D. Insulin regulation of gene expression through the forkhead transcription factor Foxo1 (Fkhr) requires kinases distinct from Akt. Biochemistry. 2001;40(39):11768–11776. doi:10.1021/bi015532m
45. Wu Y, Pan Q, Yan H, et al. Novel mechanism of Foxo1 phosphorylation in glucagon signaling in control of glucose homeostasis. Diabetes. 2018;67(11):2167–2182. doi:10.2337/db18-0674
46. Rajan MR, Nyman E, Kjølhede P, Cedersund G, Strålfors P. Systems-wide experimental and modeling analysis of insulin signaling through forkhead box protein O1 (FOXO1) in human adipocytes, normally and in type 2 diabetes. J Biol Chem. 2016;291(30):15806–15819. doi:10.1074/jbc.M116.715763
47. Armoni M, Harel C, Karni S, et al. FOXO1 represses peroxisome proliferator-activated receptor-γ1 and -γ2 gene promoters in primary adipocytes. J Biol Chem. 2006;281(29):19881–19891. doi:10.1074/jbc.M600320200
48. Zhao X, Gan L, Pan H, et al. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J. 2004;378(3):839–849. doi:10.1042/bj20031450
49. Kitamura YI, Kitamura T, Kruse J-P, et al. FoxO1 protects against pancreatic β cell failure through NeuroD and MafA induction. Cell Metab. 2005;2(3):153–163. doi:10.1016/j.cmet.2005.08.004
50. Brownawell AM, Kops GJPL, Macara IG, Burgering BMT. Inhibition of nuclear import by protein Kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol Cell Biol. 2001;21(10):3534–3546. doi:10.1128/MCB.21.10.3534-3546.2001
51. Tzivion G, Shen YH, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene. 2001;20(44):6331–6338. doi:10.1038/sj.onc.1204777
52. Rena G, Prescott AR, Guo S, Cohen P, Unterman TG. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem J. 2001;354(3):605–612. doi:10.1042/bj3540605
53. Cahill CM, Tzivion G, Nasrin N, et al. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276(16):13402–13410. doi:10.1074/jbc.M010042200
54. Wang Q, Sheng X, Shi A, et al. β-glucans: relationships between modification, conformation and functional activities. Molecules. 2017;22(2):257. doi:10.3390/molecules22020257
55. Xie Y, Li X, Ge J. Cyclophilin A–FoxO1 signaling pathway in endothelial cell apoptosis. Cell Signal. 2019;61:57–65. doi:10.1016/j.cellsig.2019.04.014
56. Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci. 2003;100(20):11285–11290. doi:10.1073/pnas.1934283100
57. Awad H, Nolette N, Hinton M, Dakshinamurti S. AMPK and FoxO1 regulate catalase expression in hypoxic pulmonary arterial smooth muscle. Pediatr Pulmonol. 2014;49(9):885–897. doi:10.1002/ppul.22919
58. Yun H, Park S, Kim MJ, et al. AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 2014;281(19):4421–4438. doi:10.1111/febs.12949
59. Zou J, Hong L, Luo C, et al. Metformin inhibits estrogen-dependent endometrial cancer cell growth by activating the AMPK-FOXO1 signal pathway. Cancer Sci. 2016;107(12):1806–1817. doi:10.1111/cas.13083
60. Yadav H, Devalaraja S, Chung ST, Rane SG. TGF-beta1/Smad3 pathway targets PP2A-AMPK-FoxO1 signaling to regulate hepatic gluconeogenesis. J Biol Chem. 2017;292(8):3420–3432. doi:10.1074/jbc.M116.764910
61. Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17(19):1646–1656. doi:10.1016/j.cub.2007.08.047
62. Lehtinen MK, Yuan Z, Boag PR, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125(5):987–1001. doi:10.1016/j.cell.2006.03.046
63. Yuan Z, Lehtinen MK, Merlo P, Villen J, Gygi S, Bonni A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem. 2009;284(17):11285–11292. doi:10.1074/jbc.M900461200
64. Maejima Y, Isobe M, Sadoshima J. Mst1 stimulates cell protective mechanisms of FoxO1 through phosphorylation. J Card Fail. 2010;16(9):S133. doi:10.1016/j.cardfail.2010.07.017
65. Maejima Y, Sadoshima J, Isobe M. Mst1 plays a cell-protective role in the heart through FoxO1 and C/EBP-β phosphorylation. J Card Fail. 2014;20(10):S147. doi:10.1016/j.cardfail.2014.07.099
66. Du X, Shi H, Li J, et al. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J Immunol. 2014;192(4):1525–1535. doi:10.4049/jimmunol.1301060
67. Kim YH, Choi J, Yang MJ, et al. A MST1–FOXO1 cascade establishes endothelial tip cell polarity and facilitates sprouting angiogenesis. Nat Commun. 2019;10(1).
68. Yuan Z, Becker EB, Merlo P, et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319(5870):1665–1668. doi:10.1126/science.1152337
69. Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314(5797):294–297. doi:10.1126/science.1130512
70. Liu P, Kao TP, Huang H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene. 2008;27(34):4733–4744. doi:10.1038/onc.2008.104
71. Lu Y, Wu Y, Feng X, et al. CDK4 deficiency promotes genomic instability and enhances Myc-driven lymphomagenesis. J Clin Invest. 2014. doi:10.1172/JCI63139
72. Ochodnicka-Mackovicova K, Bahjat M, Bloedjes TA, et al. NF-κB and AKT signaling prevent DNA damage in transformed pre-B cells by suppressing RAG1/2 expression and activity. Blood. 2015;126(11):1324–1335. doi:10.1182/blood-2015-01-621623
73. Woods YL, Rena G, Morrice N, et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355(3):597–607. doi:10.1042/bj3550597
74. Ohshima J, Wang Q, Fitzsimonds ZR, et al. Streptococcus gordoniiprograms epithelial cells to resist ZEB2 induction by Porphyromonas gingivalis. Proc Natl Acad Sci. 2019;116(17):8544–8553. doi:10.1073/pnas.1900101116
75. Wakamatsu N, Yamada Y, Yamada K, et al. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet. 2001;27(4):369–370. doi:10.1038/86860
76. Kim S, Kim Y, Lee J, Chung J. Regulation of FOXO1 by TAK1-nemo-like kinase pathway. J Biol Chem. 2010;285(11):8122–8129. doi:10.1074/jbc.M110.101824
77. Asada S, Daitoku H, Matsuzaki H, et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal. 2007;19(3):519–527. doi:10.1016/j.cellsig.2006.08.015
78. Iyer NG, Özdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23(24):4225–4231. doi:10.1038/sj.onc.1207118
79. Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi:10.1038/nature07861
80. Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101(27):10042–10047. doi:10.1073/pnas.0400593101
81. Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102(32):11278–11283. doi:10.1073/pnas.0502738102
82. Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol Endocrinol. 2005;19(9):2283–2298. doi:10.1210/me.2004-0292
83. Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi:10.1016/S0092-8674(04)00126-6
84. Masui K, Tanaka K, Akhavan D, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726–739. doi:10.1016/j.cmet.2013.09.013
85. Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285(35):27396–27401. doi:10.1074/jbc.M110.140228
86. Banks AS, Kon N, Knight C, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8(4):333–341. doi:10.1016/j.cmet.2008.08.014
87. Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280(21):20589–20595. doi:10.1074/jbc.M412357200
88. Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6(2):105–114. doi:10.1016/j.cmet.2007.07.003
89. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107(12):1470–1482. doi:10.1161/CIRCRESAHA.110.227371
90. Nakae J, Cao Y, Hakuno F, et al. Novel repressor regulates insulin sensitivity through interaction with Foxo1. EMBO J. 2012;31(10):2275–2295. doi:10.1038/emboj.2012.97
91. Choi D, Oh KJ, Han HS, et al. Protein arginine methyltransferase 1 regulates hepatic glucose production in a FoxO1-dependent manner. Hepatology. 2012;56(4):1546–1556. doi:10.1002/hep.25809
92. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi:10.1016/j.molcel.2008.12.013
93. Takahashi Y, Daitoku H, Hirota K, et al. Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 2011;13(5):505–516. doi:10.1016/j.cmet.2011.03.017
94. Yamagata K, Daitoku H, Takahashi Y, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32(2):221–231. doi:10.1016/j.molcel.2008.09.013
95. Huang J, Dorsey J, Chuikov S, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285(13):9636–9641. doi:10.1074/jbc.M109.062588
96. Jiang Z, Xing B, Feng Z, Ma J, Ma X, Hua X. Menin upregulates FOXO1 protein stability by repressing Skp2-mediated degradation in β cells. Pancreas. 2019;48(2):267–274. doi:10.1097/MPA.0000000000001239
97. Huang H, Regan KM, Wang F, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102(5):1649–1654. doi:10.1073/pnas.0406789102
98. Fu W, Ma Q, Chen L, et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284(21):13987–14000. doi:10.1074/jbc.M901758200
99. Hall JA, Tabata M, Rodgers JT, Puigserver P. USP7 attenuates hepatic gluconeogenesis through modulation of FoxO1 gene promoter occupancy. Mol Endocrinol. 2014;28(6):912–924. doi:10.1210/me.2013-1420
100. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18(7):452–465. doi:10.1038/nrm.2017.22
101. Zhang X, Qiao Y, Wu Q, et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun. 2017;8:15280. doi:10.1038/ncomms15280
102. Peternelj TT, Marsh SA, Morais C, et al. O-GlcNAc protein modification in C2C12 myoblasts exposed to oxidative stress indicates parallels with endogenous antioxidant defense. Biochem Cell Biol. 2015;93(1):63–73. doi:10.1139/bcb-2014-0106
103. Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie. 2008;90(5):679–685. doi:10.1016/j.biochi.2008.03.005
104. Housley MP, Rodgers JT, Udeshi ND, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283(24):16283–16292. doi:10.1074/jbc.M802240200
105. Cifarelli V, Lee S, Kim DH, et al. FOXO1 mediates the autocrine effect of endothelin-1 on endothelial cell survival. Mol Endocrinol. 2012;26(7):1213–1224. doi:10.1210/me.2011-1276
106. Fardini Y, Masson E, Boudah O, et al. O-GlcNAcylation of FoxO1 in pancreatic beta cells promotes Akt inhibition through an IGFBP1-mediated autocrine mechanism. FASEB J. 2014;28(2):1010–1021. doi:10.1096/fj.13-238378
107. Fardini Y, Perez-Cervera Y, Camoin L, Pagesy P, Lefebvre T, Issad T. Regulatory O-GlcNAcylation sites on FoxO1 are yet to be identified. Biochem Biophys Res Commun. 2015;462(2):151–158. doi:10.1016/j.bbrc.2015.04.114
108. Housley MP, Udeshi ND, Rodgers JT, et al. A PGC-1α-O-GlcNAc transferase complex regulates foxo transcription factor activity in response to glucose. J Biol Chem. 2009;284(8):5148–5157. doi:10.1074/jbc.M808890200
109. Shan S, Chatterjee A, Qiu Y, Hammes H-P, Wieland T, Feng Y. O-GlcNAcylation of FoxO1 mediates nucleoside diphosphate kinase B deficiency induced endothelial damage. Sci Rep. 2018;8(1). doi:10.1038/s41598-018-28892-y
110. Chen C-A, Wang T-Y, Varadharaj S, et al. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468(7327):1115–1118. doi:10.1038/nature09599
111. Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7(6):313–324. doi:10.1124/mi.7.6.7
112. Tsitsipatis D, Landrock MF, Gopal K, Klotz L-O. Modulation of FoxO1a activity through S-glutathionylation? Free Radic Biol Med. 2015;86:S34. doi:10.1016/j.freeradbiomed.2015.07.119
113. Yang J-B, Zhao Z-B, Liu Q-Z, et al. FoxO1 is a regulator of MHC-II expression and anti-tumor effect of tumor-associated macrophages. Oncogene. 2017;37(9):1192–1204. doi:10.1038/s41388-017-0048-4
114. Fan W, Imamura T, Sonoda N, et al. FOXO1 transrepresses peroxisome proliferator-activated receptor γ transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J Biol Chem. 2009;284(18):12188–12197. doi:10.1074/jbc.M808915200
115. Nakae J, Oki M, Cao Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008;582(1):54–67. doi:10.1016/j.febslet.2007.11.025
116. Ni YG, Berenji K, Wang N, et al. foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114(11):1159–1168. doi:10.1161/CIRCULATIONAHA.106.637124
117. Calissi G, Lam EWF, Link W. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov. 2020;20(1):21–38.
118. Niedan S, Kauer M, Aryee DNT, et al. Suppression of FOXO1 is responsible for a growth regulatory repressive transcriptional sub-signature of EWS-FLI1 in Ewing sarcoma. Oncogene. 2013;33(30):3927–3938. doi:10.1038/onc.2013.361
119. Kodani N, Nakae J, Kobayashi M, Kikuchi O, Kitamura T, Itoh H. FCoR-Foxo1 axis regulates α-cell mass through repression of arx expression. iScience. 2020;23(1):100798. doi:10.1016/j.isci.2019.100798
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.