Back to Journals » Infection and Drug Resistance » Volume 14
First Identification of a Patient with Prosthesis-Related Infection Caused by an MCR-1.1-Producing ST131 Escherichia coli After Rhinoplasty
Authors Li Z, Shi R, Wu H, Yan P
Received 4 December 2020
Accepted for publication 12 January 2021
Published 26 January 2021 Volume 2021:14 Pages 249—257
DOI https://doi.org/10.2147/IDR.S295801
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhehao Li,1,* Ran Shi,1,* Hao Wu,2,* Ping Yan3
1Department of Plastic and Reconstructive Surgery, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China; 2Department of Otolaryngology, Hospital of Honghe State Affiliated to Kunming Medical University, Southern Central Hospital of Yunnan Province, Mengzi, People’s Republic of China; 3Emergency Department, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ping Yan
The First Affiliated Hospital of Guangzhou Medical University, 151 Yanjiang Xi Road, Guangzhou 510120, People’s Republic of China
Tel +86 020 83336797
Email [email protected]
Background: The most common procedure of rhinoplasty is the implantation of a synthetic prosthesis. However, the complications, especially postoperative infection, could lead the suboptimal aesthetic outcome, economic losses and health threats. There is currently little literature providing an incidence of rhinoplasty infection and microbiological and antimicrobial resistance situations.
Methods: Therefore, we performed a retrospective observational study which included 173 patients who received a rhinoplasty from 1 January 2015, to 31 December 2019, in the department of plastic surgery of a tertiary hospital in Guangzhou, China. The samples from the infection site were collected and performed the bacterial culture. The antimicrobial susceptibility testing was performed by VITEK and minimum inhibition concentration testing. The whole-genome sequencing was performed by Illumina Hiseq4000 platform.
Results: We found that eight (4.6%) patients were infected by S. aureus (6), E. raffinosus (1) and E. coli (1), of which are susceptible to most antimicrobials. Remarkably, E. coli RS1231 was resistant to colistin and polymyxin B which conferred by mcr-1.1 locating on an IncI2 plasmid with 59,170-bp sequence length. Through sequence comparison, we speculate that the pRS1231S-MCR-1 was derived from animal sources. Besides, E. coli RS1231 belongs to ST131 O25:H4-fimH22 pandemic subclone and phylogroup B2, which can induce a broad variety of infections.
Conclusion: Our study provided a rhinoplasty infection incidence, microbiological and antimicrobial resistance prevalence data, and revealed, to our knowledge, the first case of postoperative infection of rhinoplasty by mcr-1.1-positive, highly susceptible, and remarkably virulent E. coli isolate.
Keywords: rhinoplasty, colistin resistance, ST131, mcr-1.1, Escherichia coli, horizontal gene transfer, virulence
Introduction
The most common procedure of rhinoplasty is the implantation of synthetic prosthesis, which is popular in East Asia. However, as a surgical procedure, postoperative complications are inevitable. The common postoperative complications include displacement, extrusion, inflammation, iatrogenic injury, scarring and infection.1 Although the infection after rhinoplasty is uncommonly reported, some processes, like osteotomies, performed during rhinoplasty breach the periosteum or nasal mucosa, creating a potential route for the spread of infection,2,3 which can lead to the failure of plastic surgery, even increase the risk of systemic infections. The basic treatment options include continued observation, antimicrobial therapy and removal of the implant; however, some therapies could influence the aesthetics of the patients, which increases the financial cost and bring the psychological blow.1,4
Staphylococcus aureus and various streptococci are the main colonization bacteria and encountered pathogens associated with postoperative infections in plastic surgery.3 To prevent postoperative infection, the use of cefazolin as a single dose is the most recommended antimicrobial for preoperative prophylaxis.4 However, the emergence of infrequent pathogens in rhinoplasty infections, especially multidrug resistance (MDR) Enterobacteriaceae, is threatening the effect of preoperative prophylaxis and the effect of antimicrobial chemotherapy.3
Based on the routine surveillance, we retrospectively observed the patients with nasal prosthesis-related infections from 1 January 2015 to 31 December 2019 in a tertiary hospital in Guangzhou, China. The results showed that the Staphylococcus aureus was the most common bacteria of the patients with postoperative infection after rhinoplasty. Specially, we found a patient, who received implantation of the synthetic prosthesis, infected with a colistin-resistant Escherichia coli isolate. Besides, we illustrated the genomic characteristics of this Escherichia coli isolate through next-generation sequencing (NGS) and confirmed mobile colistin resistance gene mcr-1.1, which was frequently reported in China with a high prevalence in food-producing animals.5
Materials and Methods
Sample Collection
For the purpose of routine monitoring of the patients with prosthesis-related infection after rhinoplasty, this study included the patients, who received rhinoplasty during a 5-year period (1 January 2015 to 31 December 2019) from the department of plastic surgery of a tertiary hospital in Guangzhou, China. The patients with rhinoplasty infection were selected for microbiological analysis. The pus samples were collected from infection site and prosthesis of these patients using nose swab and sent to the department of clinical laboratory for bacterial culture, species identification and antimicrobial susceptibility testing. Oral consent for the inclusion of research analysis for the patients was obtained.
Bacteria Isolation and Species Identification
The samples were plated on blood agar and incubated overnight at 37°C. Three of the culturable isolates were randomly selected and then subjected to species identification using VITEK 2.0 system (bioMérieux, Durham NC, USA) based on the analytical profile index. One of the isolates was subsequently subjected to the species identification verifying by 16S rDNA sequencing with PCR primers (27F: 5ʹ AGAGTTTGATCCTGGCTCAG 3ʹ; 1429R: 5ʹ GGTTACCTTGTTACGACTT 3ʹ). The species of E. coli RS1231S isolate was reconfirmed by MALDI-TOF (API 20E, bioMérieux, France).
Antimicrobial Susceptibility Testing (AST)
The obtained isolates were subjected to antimicrobial susceptibility testing using VITEK 2.0 system (bioMérieux, Durham NC, USA) using AST N-335 for gram-negative isolates (amikacin, cefepime, cefoperazone/sulbactam, ceftazidime, colistin, doxycycline, minocycline, meropenem, ciprofloxacin, levofloxacin, imipenem, tigecycline, piperacillin/tazobactam, ticarcillin clavulanic acid, aztreonam, tobramycin, trimethoprim/sulfamethoxazole) and AST GP67 (ampicillin, oxacillin, ciprofloxacin, clindamycin, gentamycin, levofloxacin, moxifloxacin, macrodantin, quinupristin/dalfopristin syncercid, rifampicin, tetracycline, trimethoprim/sulfamethoxazole, vancomycin, erythromycin, linezolid, tigecycline) for gram-positive isolates. To confirm the AST of RS1231S isolates, we performed minimum inhibitory concentrations (MICs) for colistin, polymyxin B, tigecycline, ampicillin, amoxicillin-clavulanate, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, ertapenem, imipenem, meropenem, fosfomycin, nitrofurantoin, and ciprofloxacin with three replications, using the agar dilution method excepted for colistin, polymyxin B and tigecycline which instead used the broth dilution method in accordance with the Clinical and Laboratory Standards Institute guidelines (CLSI, 2019). E. coli ATCC 25,922 was used as the standard reference strain. We interpreted the results in accordance with the CLSI breakpoints (CLSI 2019).
Next-Generation Sequencing
DNA of E. coli RS1231S was extracted using PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Genome library was prepared with 150-bp paired-end-read format and sequenced using Illumina HiSeq 4000 instrument (Illumina, Inc., San Diego, CA, United States) by Biomarker Technologies company. Sequencing data of raw sequence reads have been deposited with the NCBI (BioProject accession: PRJNA608869).
Bioinformatics Analysis
Raw reads were qualified using fastqc, trimmed and filtered with Trimmomatic to remove primer, adaptor and low-quality reads.6,7 De novo genome assembly was performed using SPAdes Genome Assembler v3.9.0.8 Assembled draft genome sequences were assessed by QUAST and annotated with RAST.9,10 The antimicrobial resistance genes (ARGs) and virulence factors (VFs) were analyzed using Blastn11 with e-value<1e-5 and identity>90%, based on comprehensive antimicrobial resistance database 3.0.7 (CARD) and virulence factor database (VFDB).12,13 The acquired ARGs and plasmid replicon type was identified by ResFinder and PlasmidFinder v2.2 was imbedded in the Center for Genomic Epidemiology (CGE; https://cge.cbs.dtu.dk/services/) with default setting.14,15 Multilocus sequence typing (MLST) of the E. coli was identified using In Silico typing tool based on the 7 housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) in Enterobase v.1.1.2.16 The mcr-1.1-harboring plasmid was closed by polymerase chain reaction (PCR) and sanger sequencing (primers: F-‘GACAGTACAGGGGCGATAC’, R-‘CGGCTGATTGTCATTTTGCG’; Annealing temperature: 59°C; Extension time: 50s; Sequence length: 370-bp). The phylogenetic tree was constructed by CSIPhylogeny using Escherichia coli str. K-12 substr. MG1655 (NC_000913.2) as the reference genome with default parameters, and visualized by iTOL.17
Results
Bacterial Prevalence of Prosthesis-Related Infection After Rhinoplasty
In this retrospective study, we included 173 patients who received rhinoplasty from 1 January 2015 to 31 December 2019. The average age of these patients was 31.2 (range: 20–51 years) and most of them are female (95.4%, 165/173). Among them, 8 (4.6%) patients had prosthesis-related infection after rhinoplasty. All these patients had local symptoms of swelling, extrusion, inflammation and pain without symptoms of systemic infection. The results of bacterial culture and species identification revealed that these eight patients were infected by several bacterial species, including S. aureus (75.0%, 6/8), E. raffinosus (12.5%, 1/8), E. coli (12.5%, 1/8) (Tables 1 and 2).
 |
Table 1 Clinical Information, Microbiological and Antimicrobial Resistance Spectrum of Patients with Gram-Positive Bacterial Infection |
 |
Table 2 Clinical Information, Microbiological, Antimicrobial Resistance and Genomic Characteristics of the Patient Infected by Colistin-Resistant E. coli RS1231S Isolate |
Antimicrobial Resistance Spectrum
We performed AST for Gram-negative and Gram-positive bacteria, respectively. More than half of S. aureus and E. raffinosus isolates were resistant to ampicillin (85.7%, 6/7), erythromycin (57.1%, 4/7) and clindamycin (42.9%, 3/7), while all were susceptible to the most antimicrobials, including tigecycline, vancomycin, linezolid. The rest of the tested antimicrobials were kept low in resistance rate (Table 1). Specially, we found that the E. coli RS1231S isolate was only resistant to colistin (MIC = 8 μg/L) (Table 2).
Epidemiological Characteristics of the Patient Infected by Colistin-Resistant E. coli RS1231S
The medical record showed that the patient infected by colistin-resistant E. coli RS1231S isolate is a 48 years old female who underwent primary nasal prosthesis implantation on 7 June 2016. Before the surgery, the patient has received cefazolin as a preoperative prophylaxis antimicrobial. The patient was admitted to the First Affiliated Hospital of Guangzhou Medical University on 28 July 2016, complaining of 5 days fever (up to 37.9°C) with redness, swelling and pain of the nose. Clinical laboratory findings on admission showed that white blood cell count was 12.31×109/mL (79.8% neutrophils granulocyte) and C-reactive protein was 31.22 mg/L, which indicated that the patient could be infected by bacterial. Then, the pus samples of the synthetic prosthesis were sampled and sent to the clinical laboratory for bacterial culture. The empirical therapy of intravenous ceftriaxone (1.0g qd) was administered for the patient, and the symptom was relieved after 5 days of treatment.
Genomic Characteristics of E. coli RS1231S Isolate
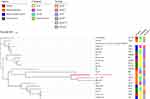
To explore the colistin resistance mechanism of E. coli RS1231S isolate, we performed NGS to investigate the genomic feature of this isolate. We obtained 20,944,110 paired-end 150-bp reads which produced a 4.98Mb genome with 66 contigs and 50.6% GC content. In silico typing revealed that E. coli RS1231S belongs to ST131 O25:H4-fimH22 pandemic subclone and phylogenetic group B2, which is commonly regarded as an infection pathogen.18,19 According to the ResFinder annotation, E. coli RS1231S harbored three acquired ARGs of mobile colistin resistance gene mcr-1.1, tetracycline resistance gene tetA and macrolides resistance gene mdfA (Table 2). Through VFDB screen, we identified 63 E. coli associated virulence genes, which involved in the categories of adherence (25.0%, 20/80), autotransporter (53.75%, 43/80), secretory system (18.75%, 15/80) and toxin (2.5%, 2/80) (Table 2). To investigate the potential sources of E. coli RS1231S isolate, we searched several genomes of MCR-1.1-producing E. coli isolate among animal, human and the environment from the publicly available database (Supplement Table 1). In the phylogenetic tree, E. coli RS1231S isolate and the most of infection-related E. coli genomes were highly correlated within a single node (Figure 1), which indicated that these infection-related isolates were genetically different from isolates from other sources.
Plasmidome of E. coli RS1231S Isolate
The plasmid replicon typing results showed that E. coli RS1231S contains six Inc types (IncFIB, IncFIC, IncI2, ColpVC, ColRNAI, ColMG828), which consistent with previous knowledge that ST131 plasmidome consisted mostly of IncF and Col-like plasmids,18,20 excepted IncI2 plasmid, indicating E. coli RS1231S belongs a pandemic subclone, which harbored various virulence factors with pathogenicity. Notably, we found that mcr-1.1 gene was located on the same contig with IncI2 replicon. Therefore, PCR and Sanger sequencing were performed to close the IncI2-mcr-1.1-harboring contig, which produced a 59,170-bp sequence as pRS1231S-MCR-1 (Figure 2). The pRS1231S-MCR-1 contained 84 coding DNA sequences (CDSs) and only one antimicrobial resistance gene mcr-1.1 was confirmed. The comparison of the first report mcr-1.1-harboring IncI2 plasmid pHNSHP45 (Accession Number KP347127.1) and pRS1231S-MCR-1 demonstrated a highly similar identity (99.99%) and coverage (92%), whereas pRS1231S-MCR-1 lost an insertion sequence IS683, an ISApl1 that located on the upstream of mcr-1.1, and a pilV associated with pilin assembly (Figure 1). Additionally, we found some loss of segments of Transposon Tn6330 (ISApl1-mcr-1.1-pap2-ISApl1) was associated with mcr-1.1 mobilization.21 However, there is no ISApl1 was found around mcr-1.1 on pRS1231S-MCR-1.
Discussion
The postoperative infection after rhinoplasty was uncommon and uncommonly reported, but can influence the suboptimal aesthetic outcome, economic losses and health threats.1 With the development of antibiotic resistance, it is important to monitor the microbiological and antimicrobial resistance for postoperative infection of rhinoplasty.3,22
In our study, we investigated the microbiological and antimicrobial resistance prevalence of 8 out of 173 patients who undergone rhinoplasty and had a postoperative infection, which is consistent with a low incidence in other countries (0–5%). Indeed, S. aureus is the most common pathogen found in rhinoplasty infections,3 whereas the E. raffinosus and E. coli were rarely reported. The antimicrobial resistance testing showed that most of S. aureus isolates and E. raffinosus isolate were resistant to ampicillin, erythromycin and clindamycin but generally susceptible to other antimicrobials, revealing the AMR situation of Gram-positive pathogen infection of rhinoplasty is not severe relatively. However, an E. coli RS1231S isolate was found in an infected patient and showed resistance to colistin, which is considered as a last-line drug to treat multi-drug resistant (MDR) Gram-negative bacteria.
For the purpose to investigating genomic characteristics of E. coli RS1231S isolate, we performed NGS and revealed that this isolate belongs to ST131 O25:H4-fimH22 pandemic subclone and phylogroup B2.20 ST131 belongs to the virulent phylogroup B2 and has been reported to induce a broad variety of infections, including meningitis, urinary tract infection, orchitis, urosepsis and peritonitis.18,19 Besides, we found 22 virulence factors associated with adherent and toxin, indicating the high ability of adhesion and pathogenicity of E. coli RS1231S isolate, which is consistent with high virulence of ST131 E. coli pandemic population. However, the infection of E. coli RS1231S isolate was susceptible to the most antimicrobials and harbored rare acquired ARGs, which is not common since E. coli ST131 is considered the quintessential example of an international multidrug-resistant high-risk clone. The susceptibility of E. coli RS1231S isolate means that there could be a good curative effect of antimicrobial treatment in this clinical case.
The plasmid-mediated colistin resistance gene mcr-1.1 was firstly reported in animals in China, which poses a challenge for MDR Gram-negative bacteria treatment.5 However, it is well investigated that food-producing animal is a reservoir of mcr-1.1.5,23–28 Besides, a low prevalence of mcr-1.1 among human infections was observed in several studies and barely reported in surgical site infections.29–31 Collectively, we speculate that the mcr-1.1-harboring IncI2 plasmid (pRS1231-S-MCR-1.1) comes from animal sources, which is primarily supported by a high coverage and an identity of comparison with animal-related plasmid sequences and a remoted genetic relation of E. coli RS1231S from other sources. The lack of ISApl1 around mcr-1.1 implying that the mcr-1.1 was stable in the plasmid sequence and unlikely transfer through transposon.21 Nevertheless, the high frequency of plasmid transfer of IncI2 plasmid needs to bring to attention.32 Even the incidence of infection by mcr-1.1-positive pathogen was rare, more surveillance should be implemented because the polymyxin was approved for the treatment of bacterial infections at the beginning of 2017 in China, which provided selective pressure in clinical infection communities. Since the development of new antimicrobials has been slow, the natural origins could be a new drug repository to explore.33–35 Since the high prevalence and spreading of mcr-1-positive pathogens are attributed to the heavy use of colistin as a feed additive in animals, the Chinese government has been officially banned the use of colistin in feed animals. The latest research showed that the mcr-1 prevalence has been dramatically decreased to a low prevalence.36 The rational use of antimicrobials should be considered as a key measure to prevent the rising of antimicrobial resistance.
In conclusion, we reported the low incidence of postoperative infection of rhinoplasty was and a relatively not severe AMR situation of these encountered pathogens. To the best of our knowledge, this is the first report of a rhinoplasty infection by mcr-1.1-positive E. coli. Future research should prospectively address the significance of AMR surveillance in the long term as well as the effectiveness of prophylactic measures and identify emerging new pathogen and resistance profile.
Ethics Statement
Written informed consent and ethics approval were exempted, since this retrospective study mainly focused on bacteria and patient intervention was not required. The patient data are confidential and compliance with the Declaration of Helsinki.
Acknowledgment
We thank Biomarker Technologies Inc. for providing assistance for the next-generation sequencing technology and bioinformatic analysis.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest for this work and declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Jang YJ, Kim SA, Alharethy S. Failure of synthetic implants: strategies and management. Facial Plast Surg. 2018;34(3):245–254. doi:10.1055/s-0038-1654676
2. Rajan GP, Fergie N, Fischer U, Romer M, Radivojevic V, Hee GK. Antibiotic prophylaxis in septorhinoplasty? A prospective, randomized study. Plast Reconstr Surg. 2005;116(7):1995–1998. doi:10.1097/01.prs.0000191181.73298.b3
3. Sharif-Askary B, Carlson AR, Van Noord MG, Marcus JR. Incidence of post-operative adverse events after rhinoplasty: a systematic review. Plast Reconstr Surg. 2019.
4. Ariyan S, Martin J, Lal A, et al. Antibiotic prophylaxis for preventing surgical-site infection in plastic surgery: an evidence-based consensus conference statement from the American Association of Plastic Surgeons. Plast Reconstr Surg. 2015;135(6):1723–1739. doi:10.1097/PRS.0000000000001265
5. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
6. Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33(19):3137–3139. doi:10.1093/bioinformatics/btx373
7. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170
8. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
9. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi:10.1093/bioinformatics/btt086
10. Overbeek R, Olson R, Pusch GD, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(D1):D206–14. doi:10.1093/nar/gkt1226
11. Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi:10.1093/nar/25.17.3389
12. Chen CY, Clark CG, Langner S, et al. Detection of antimicrobial resistance using proteomics and the comprehensive antibiotic resistance database: a case study. Proteomics Clin Appl. 2019;e1800182.
13. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi:10.1093/nar/gky1080
14. Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4(2):e27943. doi:10.4161/bact.27943
15. Carattoli A, Hasman H. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in Whole-Genome Sequencing (WGS). Methods Mol Biol. 2020;2075:285–294.
16. Zhou Z, Alikhan NF, Mohamed K, Fan Y, Agama Study G, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30(1):138–152. doi:10.1101/gr.251678.119
17. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi:10.1093/nar/gkz239
18. Price LB, Johnson JR, Aziz M, et al. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio. 2013;4(6):e00377–13. doi:10.1128/mBio.00377-13
19. Lopez-Cerero L, Navarro MD, Bellido M, et al. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J Antimicrob Chemother. 2014;69(3):809–814. doi:10.1093/jac/dkt405
20. Reid CJ, McKinnon J, Djordjevic SP. Clonal ST131-H22 Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids. Microb Genom. 2019;5(9).
21. Snesrud E, McGann P, Chandler M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio. 2018;9(1). doi:10.1128/mBio.02381-17
22. Papathanasiou M, Pohl J, Janosi RA, et al. Colonization with multiresistant bacteria: impact on ventricular assist device patients. Ann Thorac Surg. 2018;105(2):557–563. doi:10.1016/j.athoracsur.2017.07.050
23. Grami R, Mansour W, Mehri W, et al. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill. 2016;21(8):30144. doi:10.2807/1560-7917.ES.2016.21.8.30144
24. Lei L, Wang Y, Schwarz S, et al. mcr-1 in Enterobacteriaceae from companion animals, Beijing, China, 2012–2016. Emerg Infect Dis. 2017;23(4):710–711. doi:10.3201/eid2304.161732
25. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. doi:10.1016/S1473-3099(16)00061-X
26. Zhang XF, Doi Y, Huang X, et al. Possible transmission of mcr-1-Harboring Escherichia coli between companion animals and human. Emerg Infect Dis. 2016;22(9):1679–1681. doi:10.3201/eid2209.160464
27. Zhang J, Wang J, Chen L, et al. Housefly (Musca domestica) and Blow Fly (Protophormia terraenovae) as vectors of bacteria carrying colistin resistance genes. Appl Environ Microbiol. 2018;84(1).
28. Shen C, Zhong -L-L, Ma F, et al. Genomic patterns and characterizations of chromosomally-encoded mcr-1 in Escherichia coli populations. Gut Pathog. 2020;12(1):55. doi:10.1186/s13099-020-00393-2
29. Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi:10.1016/S1473-3099(16)30527-8
30. Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi:10.1016/S1473-3099(16)30528-X
31. Zhang J, Chen L, Wang J, et al. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Res Notes. 2018;11(1):143. doi:10.1186/s13104-018-3255-3
32. Meinersmann RJ. The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid. 2019;106:102444. doi:10.1016/j.plasmid.2019.102444
33. Donadu M, Usai D, Pinna A, et al. In vitro activity of hybrid lavender essential oils against multidrug resistant strains of Pseudomonas aeruginosa. J Infect Dev Ctries. 2018;12(1):9–14. doi:10.3855/jidc.9920
34. Le NT, Donadu MG, Ho DV, et al. Biological activities of essential oil extracted from leaves of Atalantia sessiflora Guillauminin Vietnam. J Infect Dev Ctries. 2020;14(9):1054–1064. doi:10.3855/jidc.12469
35. Trong N, Le D, Ho V, et al. In vitro antimicrobial activity of essential oil extracted from leaves of Leoheo domatiophorus Chaowasku, D.T. Ngo and H.T. Le in Vietnam. Plants (Basel). 2020;9(4).
36. Shen C, Zhong -L-L, Yang Y, et al. Dynamics of mcr-1 prevalence and mcr-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: a prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe. 2020;1(1):e34–e43. doi:10.1016/S2666-5247(20)30005-7
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.