Back to Journals » Infection and Drug Resistance » Volume 11
Fecal carriage and phylo-diversity of community-acquired blaTEM Enteric bacilli in Southwest Nigeria
Authors Akinduti PA , Olasehinde GI , Ejilude O, Taiwo OS, Obafemi YD
Received 26 June 2018
Accepted for publication 30 August 2018
Published 27 November 2018 Volume 2018:11 Pages 2425—2433
DOI https://doi.org/10.2147/IDR.S178243
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Paul Akinniyi Akinduti,1 Grace Iyabo Olasehinde,1 Oluwaseun Ejilude,2 Olugbenga Samson Taiwo,1 Yemisi Dorcas Obafemi1
1Microbiology Unit, Department of Biological Sciences, Covenant University, Otta, Nigeria; 2Microbiology Unit, Sacred Heart Hospital, Lantoro, Abeokuta, Nigeria
Purpose: Increasing rates of clonal spread of fecal blaTEM bacilli remains a huge concern to the community health with resultant high morbidity. The fecal carriage and clonal diversity of blaTEM within the communities in Southwest Nigeria were surveyed.
Materials and methods: Enteric bacilli obtained from fresh fecal samples randomly collected from community residents were biotyped and profiled for antibiotic susceptibility. Resistant strains were typed for beta-lactamase, extended-spectrum beta-lactamases (ESBL), AmpC and carbapenemase production while the R-plasmid carriage was detected and mating activities were examined. The presence of blaTEM gene was assayed by PCR and its phylodiversity determined with 16sRNA genomic profiling.
Results: Escherichia coli have the highest (28.6%) occurrence rate and Pseudomonas aeruginosa (20.5%) showing significant resistance to beta-lactamase inhibitors (ampicillin, cefuroxime and cefotaxime), and high-level multidrug resistance of more than 15.2% rate to ampicillin, cefuroxime, ceftazidime, tetracycline and imipenem. E. coli and Klebsiella oxytoca, are the highest beta-lactamase, ESBL and AmpC producers encoded with high molecular weight R-plasmid (>11.0 kbp) and significant rate of conjugation and transformational activities. Only 2/14, 1/13 and 1/6 ESBL-type of E. coli, K. oxytoca and Enterobacter cloaca, expressed blaTEM gene, clustering into five different phylodiverse groups with close genomic relatedness with other bacilli.
Conclusion: This is an indication of clonal dissemination of ESBL blaTEM encoded enteric bacilli having high phylodiverse characteristics through fecal carriage in the Nigerian community which requires public health education, food and environmental hygiene for its prevention.
Keywords: fecal, antibiotics, ESBL, R-plasmid, blaTEM
Introduction
Increasing rates of clonal spread of beta-lactamase enteric bacilli remains a huge concern to the community health with resultant high morbidity.1 Fecal colonization rates with extended-spectrum beta-lactamases (ESBL)-producing isolates are becoming prevalent in most communities with poor detection making implementing epidemiological measures to curtail and control the spread remain unachievable. Nevertheless, the rate of fecal carriage among the community residents remains a public health problem which has not been addressed in a few national studies.
Fecal colonization with ESBL enteric bacilli is a risk factor for acquisition of severe gastrointestinal infection and a potential source of transmission among households.2,3 With the uncontrolled use of antibiotics in most communities in Nigeria, acquisition of resistant strains considerably accelerates the selection pressure for diversification and dissemination of mutant ESBL.4,5 Currently, the high prevalence of all bla genes in different communities is aided by continuous horizontal transfer of bla-encoded plasmids among clonally unrelated strains and also through local or international transfer which played a significant role in the persistence and spread of blaTEM which was not commonly found.6,7 A number of TEM derivatives have been found to reduce affinity for beta-lactamase inhibitors showing negligible hydrolytic activity against the extended-spectrum cephalosporins.8 However, interesting mutants of TEM beta-lactamases are now being recovered and could maintain the ability to hydrolyze third-generation cephalosporins which also demonstrate inhibitor resistance called complex mutants of TEM. Plasmid-mediated horizontal transfer of blaTEM-1 genes has been demonstrated between poultry and humans with high conjugative mobility and transformational characteristics.8,9
Furthermore, the increasing rates of ESBL-expression among Gram-negative bacilli characterized with multiple clonal strains have risen substantially in several communities causing focal outbreaks resulting in high mortality rate mostly in under-developed communities having sporadic dissemination.10 Several infection-control measures focused on reducing this spread and deadly impact of its clonal transmission yielded little or no impact in many developing countries as a result of persistent spread of high level multi-antibiotic resistant strain.11 Recent mutational changes resulting in phylo-diverse beta-lactamase strains are an indication of antibiotic overuse,12 prolonged intestinal carriage13 and competitive interactions of various enteric strains within a host.14 The available therapeutic options for the treatment of ESBL-associated enteric infections are limited due to drug resistance conferred by the blaTEM, along with frequently observed co-resistance to various antibiotic classes, such as cephamycins, fluoroquinolones and aminoglycosides. A shift in the distribution of different ESBL types15 that show resistance to beta-lactam antibiotics among Gram-negative bacilli, usually confer resistance to carbapen ems and are also known to emerge as metallo-beta-lactamases (MBL).16
The recent trend in the prevalence of enteric pathogens producing blaTEM is associated with high rates of morbidity and health care costs.2 TEM gene mutations, which commonly spread with R-plasmids in enteric bacilli, sporadically reduce antibiotic efficacy.16 Therefore, this study was carried out to survey the fecal dissemination of blaTEM encoded enteric bacilli with diverse phylogenetic characteristic within the studied community.
Materials and methods
Sampling
Fresh fecal samples were randomly collected from 406 community residents who were attending the out-patient clinics. The fecal samples were transported and stored at 4°C in cold chain before analysis. For this study, approval was obtained from the Ethics committee of the Federal Medical Center, Abeokuta which also serves as referral center in Southwest, Nigeria (Ethical assign number: FMCA/470/HREC/02/2016) and also written informed consent from the individuals recruited for the study as they visited the out-patient clinics.
Culture and biotyping
Enteric bacilli were isolated from fecal samples cultured on MacConkey Agar (Oxoid CM 516, UK) and Salmonella-Shigella agar (Oxoid CM 543, UK), after incubation at 37°C for 18–24 hours. The enteric bacilli were selected according to their colony and cellular morphology and were biochemically characterized with API 24E test kit and interpreted using the manual for laboratory investigation of acute enteric infections.17
Antimicrobial susceptibility testing
Susceptibility of the enteric bacteria isolates to various commonly used antibiotics was determined using disc diffusion assay18 with the following panel; ampicillin (10 µg), amoxicillin/clavulanic acid (20 µg/10 µg), cefotaxime (30 µg), ceftazidime (30 µg), imipenem (10 µg), cefuroxime (30 µg), gentamicin (10 µg), ciprofloxacin (5 µg), tetracycline (30 µg) and azithromycin (5 µg). Briefly, 0.5 McFarlan turbid broth of pure isolate was spread on Mueller Hinton agar and each antibiotic disc was placed at a distance of about 20 mm from each other on the inoculated agar and incubated at 37°C for 18–24 hours. The inhibition zones were measured and interpreted.19
Minimum inhibitory concentration (MIC)
Resistant enteric bacilli were selected and their respective antibiotics inhibitory concentration was determined using the standard micro-dilution broth assay20 and the respective MIC of antibiotics to each isolate was determined for identification of multidrug resistant (MDR) enteric bacilli.
Phenotypic assay for beta-lactamase and ESBL production
All MDR strains were tested for the production of beta-lactamase using a modified method of starch-iodide paper acidometric method.21 The overnight broth culture of beta-lactamase producing isolates was assayed for ESBL types using the double disc method using ceftazidime (30 μg), cefotaxime (30 μg) discs, and amoxicillin-clavulanate (20/10 μg).22
Inhibitor based test for AmpC determination and Hodge test
All MDR isolates were tested for AmpC beta-lactamase inhibition on cefoxitin (30 µg) disc alone and cefoxitin (30 µg) containing boronic acid (400 µg).23 Production of carbapenemase by each strain was determined with imipenem disc (10 µg) according to the modified Hodge test (MHT) method.24
R-Plasmid profiling and mating analysis
Carriage of extra-chromosomal resistant plasmid (R-Plasmid) was investigated and profiled with alkaline lysis method25 and photo-documented with ultraviolet light to determine the DNA band sizes. Conjugative activities of the ESBL types was confirmed by mating Escherichia coli recipient strains, ATCC 25567 (penicillin-susceptible and lactose fermentation-positive strain) 1:10 ratio (donor to recipient) in 0.5% sucrose-enriched Luria-Bertani broth at 37°C, incubated for 3 hours with constant shaking and later sub-cultured on MacConkey agar plates supplemented with 6 mg/mL ceftazidime (Oxoid). Transconjugants growing in the selection plates were subjected to double disc diffusion test method26 to confirm the expression of ESBL. For transformation analysis, plasmid DNA extracted from ESBL types with alkaline lysis method was used to transform cephalosporin susceptible E. coli ATCC 25922 isolates. Briefly, 20 µL extracted DNA plasmid obtained from the ESBL isolates was added to 5 mL overnight enriched Luria-Bertani broth culture of E. coli recipient strains, ATCC 25567 (ESBL-negative and lactose fermentation-positive strain) containing 2M calcium chloride at 37°C for 6 hours.27 The mixture was later sub-cultured on selection plate of Mueller Hinton agar supplemented with 4 mg/L ceftazidime and incubated at 37°C for 18–24 hours. Transformants growing in the selection plates were subjected to double disc diffusion test method26 prescribed by Apfalter et al., (2007) to confirm the presence of ESBL resistance determinants present in the recipient isolates.
PCR assays for TEM beta-lactamase genes
Chromosomal DNA from ESBL strains were extracted by boiling colonies of bacteria emulsified in RNAse-free water at 55°C for 10 minutes, centrifuged at 12,000 rpm for 5 minutes and the supernatant was carefully removed into another clean tube as DNA template for amplification. The extracted chromosomal DNA was adjusted using Thermoscientific Nanodrop Spectrophotometer of absorbance of 260 nm filter before adding to PCR mix. Each PCR assay was carried out in a volume of 2.5 µL 10× PCR buffer, 1.5 µL 25 mM MgCl2, 0.5 µL of 10 mM dNTPs (dATP, dGTP, dCTP, dTTP), 0.2 µL Forward primer of 2500 pmole (5-AGATCCGATCATGAGACAATA-3), 0.2 µL of Reverse primer of 2500 pmole (5-TCTAGGCTAG TACTCTGTTAT-3), 0.2 µL Taq polymerase enzymes, 17.9 µL distilled water and 2.0 µL of each extracted chromosomal DNA to make final reaction volume of 25.0 µL and amplified accordingly (Table 1).
  | Table 1 PCR conditions used for the detection of TEM beta-lactamase genes |
Genotypic characterization of TEM beta-lactamase strains
Extraction of chromosomal DNA of each ESBL strain was carried out using QIAamp DNA Mini Kit (Qiagen, USA) and estimated on Thermoscientific Nanodrop Spectrophotometer at an absorbance of 260 nm filter. Amplification for the 16sRNA assay was carried out in a volume of 25 µL reaction containing 2.5 µL of 10× Bioline Taq reaction buffer, 2 µL of 2.5 mM dNTPs (dATP, dGTP, dCTP, dTTP), 1 µL of 5 µM Forward primer 16sRNA-U1 (CCAGCAGCCGCGG AATACG), 1 µL of 5 µM Reverse primer, 16sRNA-U2 (ATC GGTACC T TG TACGACTTC),28 10 ng/µL DNA template of 3 µL, 0.4 µL DNA Taq polymerase (5 unit/μL) and nuclease-free water to make up to the volume of 25 μL. The amplification reaction was carried out in 32 repeated temperature cycles in a thermal cycler block (ATC 401; Nyx Technik) at a temperature of 94°C for 5 minutes, denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds and extension at 72°C for 45 seconds.
Amplicon sequencing and BLAST
Amplicon products were purified with QIAquick PCR purification kit (Qiagen, USA) and 6 µL diluted DNA amplicon was added to 4 µL of 16sRNA primer at a concentration of 0.8 pmol/µL and the reaction mixtures were added to Taq sequencing preparations (containing tag-oligonucleotide Taq polymerase) according to the manufacturer (Applied Biosystem Company, USA) instructions and sequenced using ABI Prism Big Dye Terminator version 3.0 cycle and the products were subsequently analyzed on an ABI PRISM 3700 DNA. The nucleotide sequences were translated for molecular characterization on GeneBank (www.ncbi.org) and more than 98% similarity was considered.
Phylogenetic analysis
Nucleotides obtained from 16sRNA sequences from ESBL type enteric bacilli were analyzed for diversity with highly similar sequences from different geographical locations having highest percentage coverage query and identity from Gen Bank database with their repository accession numbers. Nearest phylodiversity was determined by manually aligning the ESBL type sequences with homologous sequences configured into multiple sequence alignment in MEGA 6 software version 629 (http://www.megasoftware.net) to construct dendrogram by Maximum Likelihood method as independent, unordered and equally weighted according to Fitch parsimony with 1,000 heuristic bootstrap replicates and substitution model as “p” distance.
Data analysis
The significance of the antibiotic profile of the isolates was analyzed with SPSS software version 20.0 using Chi-square to evaluate the differences between susceptibility, intermediate and resistance level of fecal bacilli and the significant differences by ANOVA for R-plasmid carriage, conjugants and transformants at P<0.05.
Results
Antibiogram of isolated bacilli
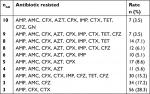
Of all the enteric isolates obtained from the community subjects, E. coli showed highest occurrence rate (28.6%), followed by Pseudomonas aeruginosa (20.5%) while Salmonella spp. and Shigella spp. were the least found isolates (Figure 1). A significant rate of 52.2%, 53% and 56% enteric bacilli showed resistant to beta-lactamase antibiotics (ampicillin, cefuroxime and cefotaxime), and low resistance to ciprofloxacin (18.8%) and gentamycin (18.2%), respectively (P=0.001) (Table 2). Majority of the enteric bacilli were observed to be MDR to ampicillin, cefuroxime, amoxicillin-clavulanic acid and azithromycin whereas more than 15.2% of resistant bacilli reveal high MDR profile to more than two classes of antibiotics such as ampicillin, cefuroxime, ceftazidime, tetracycline and imipenem (Table 3).
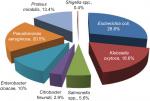  | Figure 1 Distribution of the enteric isolates obtained from the community people (n=479). |
  | Table 2 Antibiotic susceptibility pattern of enteric bacilli (N=479) Note: Significant P<0.05. |
Beta-lactamase production, ESBL-types and mating activities of resistant enteric bacilli
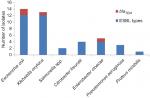
Comparative distribution of beta-lactamase producers were highest among E.coli and Klebsiella oxytoca, and the similar high rate was observed in ESBL assay while AmpC and carbapenamase production were low and no Salmonella spp. produce AmpC beta-lactamase and carbapenamase (Figure 2). In Table 4, the highest rate of 52.6% harbored high molecular weight R-plasmid (>11.0 kbp) and 2.6% express low molecular weight R-plasmid (<5 kbp) while 21.1% possessed no R-plasmid. Mating activity revealed the significant rate of 29% and 34.4% of the isolates having high plasmid weight to conjugant and transform recipient isolates effectively (P<0.05).
  | Figure 2 Comparative distribution of beta-lactamase, ESBL, AmpC and carbapenamase types among the multi-antibiotics resistant isolates. Abbreviation: ESBL, extended-spectrum beta-lactamases. |
Genotyping and clonal diversity of blaTEM isolates
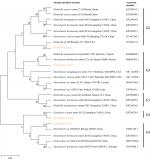
Genomic amplification of blaTEM genes recovered from ESBL type (Figure 3) showed 2/14, 1/13 and 1/6 of E. coli, K. oxytoca and Enterobacter cloaca expressing blaTEM (Figure 4), respectively. Clonal diversity of the 16sRNA gene sequence of ESBL-types carrying blaTEM gene recovered from community residents in Southwest Nigeria revealed cluster into five diverse phylogroups (G) with other bacilli deposited in GenBank. E. coli F1 from Nigerian community clustered with Enterobacter spp. from China, India and Portugal into phylogroup 4 (G4). High-level phylodiversity was shown by E. coli F2 clustering to G3 with Klebsiella michiganensis found in Port-Harcourt, Nigeria and Enterobacter hormaechei (Mexico) while similar clonal diversity was expressed by K. oxytoca clustered together with seven other bacilli in G2 but closely related to Klebsiella spp. and phylogroup G5 clustered E. cloaca from community subjects with Kosakonia cowanii from China (Figure 5).
  | Figure 3 blaTEM genes recovered from ESBL type isolates on agarose gel. Abbreviation: ESBL, extended-spectrum beta-lactamases. |
  | Figure 4 Clonal distribution of blaTEM genes among ESBL types among the enteric bacilli. Abbreviation: ESBL, extended-spectrum beta-lactamases. |
Discussion
Investigation of disseminating ESBL types is now a recognized issue in emerging global health threat due to unfavorable treatment outcome of common enteric infectious diseases particularly in several communities. High occurrence rates of MDR enteric bacilli (E. coli, Pseudomonas aeruginosa, Salmonella spp. and Shigella spp.) to beta-lactamase antibiotics (ampicillin, cefuroxime and cefotaxime) suggest an increasing prevalence of resistant strains which is gradually becoming a nightmare and a fatal challenge for the community health.30 Unfortunately, reports regarding community bacterial resistance to antimicrobial agents are limited in Southwest Nigeria. Therefore, the increasing trends and a recorded upsurge in resistance and prevalence of community-acquired ESBL-types is not yet clear, but associations with foodstuffs, consumption of animals fed with antibiotics and frequent contact with patients during their illness could be major factors to consider.
Comparatively, high distribution of E. coli and K. oxytoca ESBL and AmpC types is a reflection of emerging community-acquired ESBL strains which could be elicited by self-medication, uncontrolled use of antibiotics and unhygienic eating habits in numerous poorly located restaurants and street food vendors in many communities.31 This high level production of AmpC beta-lactamases and carbapenamase by community E. coli, K. oxytoca and Citrobacter freundii poses a grave danger to the residents.32,33 Furthermore, increased use of broad-spectrum antibiotics during prolonged infections usually aids resistance with severe complications and financial loss.
Fecal carriage of high molecular weight R-plasmid (>11.0 kbp) among ESBL types correlated with plasmids encoding resistance to other antibiotics such as aminoglycosides, sulfonamides, tetracyclines and other antibiotics.34,35 The unforeseen trend in plasmid-mediated resistance via heavy plasmid coding for ESBL carriage and intestinal colonization of MDR strains to other class of antibiotics may mask the spread of these clones among the residents. The plasmid-mediated ESBL mainly confer resistance on oxyimino-β-lactams, such as cefotaxime, ceftazidime, ceftriaxone and aztreonam, causing selective pressure for antibiotic resistant transfer to other isolates. Conjugative property of the ESBL bacilli further confirms the role of food hawked in the community as a potential reservoir of antibiotic resistance bacteria that contained a pool of mobile genetic elements (plasmid), which are readily disseminated to other human pathogens.36 Having high degree of transformation activity, identified ESBL types could propagate spread of resistant genes at high frequency that could be promoted by constant release of naked DNA from dying bacilli which can be taken up by other isolates, integrating into the cell genome and enhancing undetected spread within the community.37
Clonal transmission of antibiotic resistance may be not only due to dissemination of different β-lactamases38 but also by spread of clonal diverse variants with high potential plasmid-mediation and mating activity. The study shows only 2 of 14, 1 of 13 and 1 of 6 ESBL-type of E. coli, K. oxytoca and E. cloaca expressed blaTEM gene, respectively. Clonal diversity of the 16sRNA gene sequence of ESBL-types carrying blaTEM gene recovered from community residents in Southwest Nigeria revealed close relatedness with other enteric pathogens from other countries. TEM-type ESBL (blaTEM gene) known to hydrolyze ampicillin at a greater rate but with less activity against extended-spectrum cephalosporins, is observed to be responsible for resistance in commonly used third generation cephalosporins in this locality.39 Expression of blaTEM gene observed in community ESBL isolates reveals resistance to ceftazidime with very high MIC values. Therefore, molecular epidemiological studies need to be carried out to prevent dissemination of this strain in order to curtail the spread of enteric infections and prevention of major outbreaks of blaTEM ESBL strains in the Nigerian community. High-level phylodiversity observed among the Southwest Nigeria isolates reveals transfer of 16sRNA genotyped bacilli harboring blaTEM gene which might have been transmitted from clones of different origins.1
Frequent household contact with pets,40 animal food41 and in-door reared birds,42 as well as horizontal transfer of gene within the commensal microflora43 could necessitate clonal diversity of the blaTEM ESBL types and also serve as vehicles for undetected clonal spread of antimicrobial-resistant bacilli.44 Consumption of poultry products such as birds treated with antibiotic or fed with antibiotic-supplemented feed, imported from neighboring countries into Southwest Nigeria could be implicated as source of blaTEM clones circulating within the community, thereby increasing intestinal colonization and resultant carriage level of blaTEM bacilli among the community residents. This poses a grave danger to the populace and the community residents, making routine surveillance of antimicrobial susceptibility difficult and formulation of public health guidelines unachievable.
Conclusion
Identification of blaTEM carriage among enteric bacilli circulating in the Nigerian community, having high phylodiverse characteristic needs concerted effort to prevent its dissemination and practicable public health education on food and environmental hygiene.
Acknowledgments
The authors appreciate the management of Covenant University, Ota, Nigeria for funding the publication of this research study and Federal Medical Centre, Abeokuta for the collection and storage of the samples. Also, the Biomedical Research Group, Abeokuta Nigeria for partly funding the study.
Disclosure
The authors report no conflicts of interest in this work.
References
Jena J, Debata NK, Sahoo RK, Gaur M, Subudhi E. Genetic diversity study of various β-lactamase-producing multidrug-resistant Escherichia coli isolates from a tertiary care hospital using ERIC-PCR. Indian J Med Res. 2017:;146(Suppl.):S23–S29. | ||
Valverde A, Grill F, Coque TM, et al. High rate of intestinal colonization with extended-spectrum-beta-lactamase-producing organisms in household contacts of infected community patients. J Clin Microbiol. 2008;46(8):2796–2799. | ||
Rodríguez-Baño J, López-Cerero L, Navarro MD, Díaz de Alba P, Pascual A. Faecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. J Antimicrob Chemother. 2008;62(5):1142–1149. | ||
Akinduti P, Oluwadun A, Iwalokun B, Onagbesan O, Ejilude O. Community-Acquire CTX-M Beta-Lactamase Enteric Isolates in Abeokuta, Nigeria. Br Microbiol Res J. 2014;5(4):351–358. | ||
Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42(1):1–17. | ||
Cloeckaert A, Praud K, Doublet B, et al. Dissemination of an extended-spectrum-beta-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob Agents Chemother. 2007;51(5):1872–1875. | ||
Novais A, Cantón R, Machado E. International dissemination of a multi-resistant IncA/C2 plasmid containing blaTEM-24, Tn21 and Tn1696 among epidemic and non-epidemic Enterobacteriaceae species. Antimicrob. Agents Chemother. 2008;41:223–232. | ||
Singh NS, Singhal N, Virdi JS. Genetic Environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and Characterization of Integrons of Escherichia coli Isolated From an Indian Urban Aquatic Environment. Front Microbiol. 2018;9:382. | ||
Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56(3):463–469. | ||
Kothari C, Gaind R, Singh LC, et al. Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 2013;13:136. | ||
Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64(Supp l):i3–i10. | ||
Jafari RA, Motamedi H, Maleki E, Ghanbarpour R, Mayahi M. Phylogenetic typing and detection of extended-spectrum β-lactamases in Escherichia coli isolates from broiler chickens in Ahvaz, Iran. Vet Res Forum. 2016;7(3):227–233. | ||
Yan JJ, Ko WC, Tsai SH, Wu HM, Jin YT, Wu JJ. Dissemination of CTX-M-3 and CMY-2 beta-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol; 2000(38):4320–4325. | ||
Gordon DM, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149(Pt 12):3575–3586. | ||
Bratu S, Landman D, Haag R, et al. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med. 2005;165(12):1430–1435. | ||
Oteo J, Navarro C, Cercenado E, et al. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J Clin Microbiol. 2006;44(7):2359–2366. | ||
World Health Organization. Manual for the laboratory investigations of acute enteric infections. 1987. Publication no. WHO/CDD/83.3 rev 1. World Health Organization, Geneva, Switzerland. | ||
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. | ||
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Nineteenth informational supplement. CLSI-M100-S22. Wayne, PA: CLSI; 2012. | ||
Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. | ||
Odugbemi TO, Hafiz S, McEntegart MG. Penicillase producing Neisseria gonorrhoea. Detection by Starch Iodide Paper Techniquies. Br Med J. 1977;11:500. | ||
Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48(Suppl 1):59–64. | ||
Coudron PE. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J Clin Microbiol. 2005;43(8):4163–4167. | ||
Landman D, Salvani JK, Bratu S, Quale J. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J Clin Microbiol. 2005;43(11):5639–5641. | ||
Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513–1523. | ||
Apfalter P, Assadian O, Daxböck F, Hirschl AM, Rotter ML, Makristathis A. Extended double disc synergy testing reveals a low prevalence of extended-spectrum beta-lactamases in Enterobacter spp. in Vienna, Austria. J Antimicrob Chemother. 2007;59(5):854–859. | ||
Smith CJ, Markowitz SM, Macrina FL. Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother. 1981;19(6):997–1003. | ||
Rahmani S, Forozandeh M, Mosavi M, Rezaee A. Detection of bacteria by amplifying the 16s RNA gene with universal primers and RFLP. Med J Islam Repub Iran. 2006;19(4):333–338. | ||
Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S. 2013. MEGA 6. Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. | ||
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. | ||
Iroha I, Ikeagwu I, Ejikeugwu C, et al. Prevalence and antibiogram of Pathogenic Gram Negative Bacteria Isolated from patients Seeking Medical Care in a Tertiary Hospital in Abakaliki, Nigeria. Adv Res J Med Med Sci. 2015;4:455–460. | ||
Shanthi M, Sekar U, Arunagiri K, Sekar B. Detection of Amp C genes encoding for beta-lactamases in Escherichia coli and Klebsiella pneumoniae. Indian J Med Microbiol. 2012;30(3):290–295. | ||
Afiukwa FN, Iroha IR, Afiukwa CA, Ayogu TE, Oji AE, Onwa NC. Presence of coliform producing extended spectrum beta lactamase in sachet-water manufactured and sold in Abakaliki, Ebonyi state. Nigeria International Research Journal Microbiology. 2010;1(2):032–036. | ||
Hemalatha V, Padma M, Sekar U, Vinodh TM, Arunkumar AS. Detection of Amp C beta lactamases production in Escherichia coli & Klebsiella by an inhibitor based method. Indian J Med Res. 2007;126(3):220–223. | ||
Motayo BO, Akinduti PA, Adeyankinu FA, et al. Antibiogram and plasmid profiling of carbapenamase and extended spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumonia in Abeokuta, South western, Nigeria. African Health Sciences 2013;13(4):1140–1146. | ||
Sharma J, Ray P, Sharma M, Jyoti S, Pallab R, Meera S. Plasmid profile of ESBL producing Gram-negative bacteria and correlation with susceptibility to beta-lactam drugs. Indian J Pathol Microbiol. 2010;53(1):83–86. | ||
Van TTH, Moutafis G, Tran LT, Coloe PJ. Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl Environ Microbiol. 2007;73(24):7906–7911. | ||
Guillet M, Bille E, Lecuyer H, et al. Epidemiology of patients harboring extended-spectrum beta-lactamase-producing enterobacteriaceae (ESBLE), on admission. Med Mal Infect. 2010;40(11):632–636. | ||
Loncaric I, Stalder GL, Mehinagic K, et al. Comparison of ESBL – and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One. 2013;8(12): e84048. | ||
Gonçalves A, Igrejas G, Radhouani H, et al. Iberian wolf as a reservoir of extended-spectrum β-lactamase-producing Escherichia coli of the TEM, SHV, and CTX-M groups. Microb Drug Resist. 2012;18(2):215–219. | ||
Nowrouzian F, Hesselmar B, Saalman R, et al. Escherichia coli in infants’ intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr Res. 2003;54(1):8–14. | ||
Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother. 2013;68(1):60–67. | ||
Ünal N, Karagöz A, Aşkar Ş, Dilik Z, Yurteri B. Extended-spectrum β-lactamases among cloacal Escherichia coliisolates in healthy broilers in Turkey. Turk J Vet Anim Sci. 2017;41:72–76. | ||
Cristóvão F, Alonso CA, Igrejas G, et al. Clonal diversity of extended-spectrum beta-lactamase producing Escherichia coli isolates in fecal samples of wild animals. FEMS Microbiol Lett. 2017;364(5):1–6. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.