Back to Journals » Infection and Drug Resistance » Volume 13
Fecal Carriage and Molecular Epidemiology of Carbapenem-Resistant Enterobacteriaceae from Inpatient Children in a Pediatric Hospital of Shanghai
Authors Xu Q, Pan F, Sun Y, Wang C , Shi Y, Zhang T, Yu F, Zhang H
Received 6 August 2020
Accepted for publication 12 November 2020
Published 10 December 2020 Volume 2020:13 Pages 4405—4415
DOI https://doi.org/10.2147/IDR.S275549
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Qi Xu, Fen Pan, Yan Sun, Chun Wang, Yingying Shi, Tiandong Zhang, Fangyuan Yu, Hong Zhang
Department of Clinical Laboratory, Shanghai Children’s Hospital, Shanghai Jiaotong University, Shanghai, People’s Republic of China
Correspondence: Hong Zhang Department of Clinical Laboratory
Shanghai Children’s Hospital, Shanghai Jiaotong University, No. 24, Lane 1400, West Beijing Road, Shanghai 200040, People’s Republic of China
Tel/ Fax +86 21 6247 4880 Ext 82301
Email [email protected]
Purpose: To determine the epidemiology characteristics of intestinal colonization of carbapenem-resistant Enterobacteriaceae (CRE) among inpatients in a pediatric hospital in China.
Methods: A retrospective study was conducted from April to December 2019. Medical records were reviewed to extract the clinical information. Antimicrobial susceptibility was performed by broth microdilution method. Drug resistance determinants and plasmid types were analyzed using polymerase chain reaction (PCR) assays. Multilocus sequence typing (MLST) and Enterobacterial repetitive intergenic consensus sequences PCR (ERIC-PCR) were employed to determine the genetic relationships between strains.
Results: A total of 90 CRE strains were isolated, with a fecal carriage rate of 8.6% (90/1052), and mainly distributed in E. aerogenes (n=30), K. pneumoniae (n=25) and E. coli (n=23). More than 50% of CRE colonizers had a history of invasive procedures and antibiotic exposures. As high as 91.1% (82/90) of CRE isolates carried carbapenemase genes, with blaNDM-5 (n=56) being the most common, and mainly found in E. aerogenes (51.8%, 29/56) and E. coli (32.1%, 18/56) isolates, which primarily belonged to ST4 (100%, 29/29) and ST692 (55.6%, 10/18), respectively. Followed by blaKPC-2 (n=12), and all found in K. pneumoniae ST11 isolates. Other carbapenemase genes including blaNDM-1, blaIMP-4 and blaIMP-26. Meanwhile, ESBL genes (blaCTX-M, blaTEM-1 and blaSHV) and AmpC genes (blaDHA-1 and blaEBC) were also detected. All CRE isolates showed high resistance to cephalosporins and carbapenemases (97.8%-100.0%) but remained susceptible to tigecycline (98.9%). IncX3 was a major plasmid type in NDM-containing strains (91.3%), and 91.7% of KPC-2-producing K. pneumoniae harboring IncFII and IncFIB plasmids. The ERIC-PCR revealed that several strains with identical STs were genetically similar.
Conclusion: This study revealed a major intestinal colonization of ST4 NDM-5 E. aerogenes, ST11 KPC-2 K. pneumoniae and ST692 NDM-5 E. coli strains among inpatients in a pediatric hospital. Infection control measures should be implemented immediately to prevent the spread of these strains in clinical settings.
Keywords: intestinal colonization, hospitalized children, ST4, E. aerogenes, NICU, ERIC-PCR
Introduction
Carbapenems used to be the most effective agents for the treatment of multi-drug resistant (MDR) bacterial infections, but the emergence and dissemination of carbapenem resistance have been increasingly reported across the world since 1990s.1,2 The global spread of carbapenem-resistant Enterobacteriaceae (CRE) is one of the most severe threats to human health in clinical settings. According to the nationwide surveillance of bacterial resistance data in China, the resistance rate of K. pneumoniae to imipenem and meropenem increased to 25% and 26.3% in 2018, respectively, from 3.0% and 2.9% in 2005.3 In Europe, the prevalence of carbapenem-resistant K. pneumoniae (CR-KPN) has risen to 60% in Greece and 40% in Italy already.4,5 The production of carbapenemases (KPC, NDM, and OXA-48-like) is the major mechanism among CRE isolates,6,7 and the rapid worldwide spread of CRE largely attributes to the dissemination of carbapenemase enzymes.8
In the last 10–15 years, CRE has become a major cause of healthcare-associated infections (HAI).9 Intestinal colonization with CRE has been considered as a significant risk factor for subsequent infection, a previous cohort study demonstrated that nearly 50% CRE-colonized patients developed a CRE infection within 30 days compared to non-colonized patients, and the odds of infection increased by 10.8 times.10 As we all know, patients with CRE colonization or infection have been associated with higher healthcare costs, prolonged hospital stays, treatment failures and mortality, as well as wide usage of broad-spectrum antimicrobial agents.11 Several studies have been conducted to investigate the epidemiology characteristics of intestinal colonization of CRE strains in hospitalized adults.12,13 However, the fecal carriage of CRE in hospitalized children has not been well studied, especially in China. Therefore, our study was conducted to determine the clinical characteristics, antimicrobial resistance profiles and molecular epidemiology of intestinal colonization of CRE among inpatients in a pediatric hospital in Shanghai, China.
Methods
Samples Collection and Strains Screening
The study was performed from April to December 2019 retrospectively in Shanghai Children’s Hospital, which is a 700-bed specialized pediatric teaching hospital, serving a population of more than 2.4 million children annually, and nearly 50,000 are inpatients. We consulted electronic medical record of each inpatient for clinical information. This study was reviewed and approved by the Ethics Committee of Shanghai Children’s Hospital in accordance with the Declaration of Helsinki and its amendments or comparable ethical standards. The patient informed consent was waived because our study only focused on the bacterial isolates and did not have an influence on the patients.
Fecal swabs were consecutively collected from hospitalized children who received the fecal culture testing, and stored in Cary-Blair transport medium (Hopebio, Qingdao, China) in 4°C condition up to 1 day prior to testing. For each patient, only the first fecal sample was considered. The CRE isolates were detected by inoculating in home-made MacConkey agar supplemented with meropenem at 1μg/mL and then incubated at 35±2°C in a 5% CO2 incubator for 24–48 h, the growth of at least one colony per agar plate was considered as a positive result. All strains screened were stored at −80°C in 40% glycerol broth medium for further analysis.
CRE Identification and Antimicrobial Susceptibility Testing
All screened strains were identified by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry using MALDI Biotyper (Bruker Daltonik GmbH, Bremen, Germany). A panel of 17 antimicrobial agents were used to determine the antimicrobial susceptibility of all CRE isolates, including cefotaxime, ceftazidime, cefoperazone, cefepime, ertapenem, imipenem, meropenem, ciprofloxacin, cefoperazone-sulbactam, piperacillin–tazobactam, amikacin, gentamicin, trimethoprim-sulfamethoxazole, aztreonam, ceftazidime-avibactam, fosfomycin, colistin and tigecycline. The minimum inhibitory concentrations (MICs) was determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) breakpoints, and E. coli strain ATCC 25922 was used for quality control. The interpretive criterion for colistin and tigecycline was based on the breakpoints of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Food and Drug Administration (FDA), respectively. MDR was defined as acquired nonsusceptibility to at least one agent in three or more antimicrobial categories.14
Detection of Resistance Genes of CRE
Carbapenemase genes (blaNDM, blaKPC, blaIMP, blaVIM, blaOXA-48, blaAIM, blaGIM, and blaSIM), ESBL genes (blaCTX-M, blaTEM and blaSHV), AmpC genes (blaMOX, blaFOX, blaDHA, blaCIT, blaAAC and blaEBC) and the colistin resistance genes (mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5) were identified by PCR assays. Primer design and amplification conditions were based on the previous reports.15–19 The amplified products were sequenced by Sangon Biotech (Shanghai) Co. Ltd., and then compared to the sequences available at the National Center for Biotechnology Information (NCBI) website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Plasmid Replicon Typing
For all CRE isolates, the plasmids were typed based on their incompatibility group using PCR-based replicon typing (PBRT), and a total of 21 pairs of primers (HI1, HI2, I1, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIAs, F, K, B/O, X1, X2, X3 and X4) were amplified as previously described.20,21 The amplified products were then electrophoresed in 1.5% agarose gels, stained with GeneGreen Nucleic Acid Gel Stain (TIANGEN, Beijing, China) and visualized under ultraviolet light using the Gel Doc 2000 system (Bio-Rad).
Multilocus Sequence Typing (MLST)
MLST was performed using the schemes hosted on the Pub MLST website (https://pubmlst.org/) and Institute Pasteur MLST website (https://bigsdb.pasteur.fr/). The housekeeping genes of E. aerogenes and E. Cloacae (dnaA, fusA, gyrB, leuS, pyrG, rplB and rpoB), K. pneumoniae (ropB, gapA, mdh, pgi, phoE, infB and tonB) and E. coli (dinB, icdA, pabB, polB, putB, trpA, trpB and uidA) were amplified and sequenced, then the STs were obtained by comparing the sequences in the MLST database.
Enterobacterial Repetitive Intergenic Consensus Sequences PCR (ERIC-PCR)
The ERIC-PCR was applied using the ERIC-F (5ʹ- ATGTAAGCTCCTGGGGATTCAC-3ʹ) and ERIC-R (5ʹ-AAGTAAGTGACTGGGGTGAGCG-3ʹ) primers for E. aerogenes, K. pneumoniae and E. coli isolates, and the amplification conditions were adjusted according to the reports published previously.22,23 The gel electrophoresis image was analyzed by BioNumerics7.6 version software (Applied Maths; NV Keistraat, Sint-Martens-Latem, Belgium). Dice coefficients were used to calculate the similarity of ERIC-PCR patterns, of which optimization set at 1% and tolerance at 1%. Dendrograms were constructed by the unweighted pair group method with arithmetic averages (UPGMA) and isolates were categorized into the same cluster with a cutoff value of 90% similarity. Identical strains were defined as isolates with 100% similarity.
Statistical Analysis
Data were described as median and interquartile ranges (25th and 75th percentile), the number of cases or percentages. The quantitative data were compared by t-test and variance analysis, categorical data were evaluated by Chi-square or Fisher’s exact test. A P-value of <0.05 was considered statistically significant, and all statistical analysis was performed using SPSS version 26.0 software (IBM, Armonk, NY). The antibiotic resistance data were analyzed with WHONET 5.6.
Results
Samples Collection and CRE Distribution
A total of 1612 consecutive and non-duplicate fecal samples were collected from inpatients between April to December in 2019, getting rid of 560 samples from patients hospitalized for less than 48 h, there were totally 90 CRE strains isolated as a carriage rate of 8.6% (90/1052), including E. aerogenes (n=30, 33.3%), K. pneumoniae (n=25, 27.8%), E. coli (n=23, 25.6%), E. cloacae (n=9), K. oxytoca (n=2) and C. freundii (n=1). The CRE strains mainly distributed in neonatal intensive care unit (NICU) (45.6%, 41/90), pediatric intensive care unit (PICU) (22.2%, 20/90) and gastroenterology ward (20.0%,18/90) (Figure 1). It is worth mentioning that, compared to E. aerogenes isolates, more K. pneumoniae strains were isolated from PICU than NICU (P<0.05), and 43.5% (10/23) and 30.4% (8/23) of E. coli isolates originated from NICU and PICU, respectively. In addition, we also found CRE isolates in respiratory, hematology, special consultation, nephrology and general surgery wards.
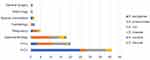 |
Figure 1 Distribution of CRE strains in different departments. Abbreviations: PICU, pediatric intensive care unit; NICU, neonatal intensive care unit. |
Clinical Characteristics
As shown in Table 1, the median age of patients colonized with CRE isolates was 2 months (interquartile range, 0.7–12 months), male to female ratio was 1.8 to 1. Compared to patients carried E. aerogenes and E. coli isolates, children who colonized with K. pneumoniae had a longer hospitalization time (P<0.05). The median length from admission to CRE detected was 12 days. Many children were accompanied by severe underlying disease such as gastrointestinal diseases (22.2%), Immunocompromised (12.2%), neonatal respiratory distress syndrome (NRDS) (12.2%) and chronic heart disease (CHD) (4.4%). More than 50% CRE colonizers had a history of invasive procedures and antibiotic exposures. In addition, the total cure and death rate were 81.1% and 6.7%, respectively.
 |
Table 1 The Clinical Characteristics of CRE Carriers |
We also retrospectively reviewed the clinical information of patients with CRE colonized in NICU (Table 2) and observed an interesting phenomenon that the birth weight of E. coli colonizers (Median, 1360 g) were lighter than the E. aerogenes colonizers (Median, 1630g) (P<0.05), and the gestational age was also younger (P<0.05), the Apgar Score at 5 min was 8. Breastfeeding was the main feeding options in newborns (68.3%), and 5 neonates at birth were also found to be colonized with CRE. During pregnancy, the diseases of pre-eclampsia (24.4%) and gestational diabetes (22.0%) were more common, and dexamethasone was the most frequently used agent (19.5%), followed by antihypertensives (12.2%) and antibiotics (9.8%). A large proportion of neonates were born via cesarean delivery (61.0%).
 |
Table 2 The Clinical Characteristics of CRE Carriers in NICU |
Carbapenemase and Other Resistance Genes
As high as 91.1% (82/90) of CRE isolates carried carbapenemase genes, with blaNDM-5 (n=56) being the predominant carbapenemase genotype. The blaNDM-5 gene was most common in E. aerogenes (51.8%, 29/56) and E. coli isolates (32.1%, 18/56), which was also observed in K. pneumoniae (n=6) and E. cloacae isolates (n=3). The second most common gene was blaKPC-2 (n=12), which was all found in K. pneumoniae isolates. In addition, 6 strains harbored blaNDM-1, including 2 E. coli, 2 E. cloacae, 1 K. oxytoca and 1 C. freundii isolates. Moreover, 2 K. pneumoniae isolates carried blaIMP-4, and 4 strains possessed both blaNDM-1 and blaIMP-4, containing K. pneumoniae (n=2), E. cloacae (n=2) and K. oxytoca (n=2) isolates, but the blaIMP-26 was only identified in E. cloacae isolates.
Meanwhile, ESBL genes such as blaCTX-M-14 (n=62), blaTEM-1 (n=61) and blaCTX-M-15 (n=51) were also detected in these CRE isolates, and blaSHV-11 and blaSHV-1 were mainly found in K. pneumoniae isolates (21/30), besides, blaSHV-12 (n=4) and blaSHV-5 (n=1) were only observed in E. cloacae isolates. What’s more, 39 CRE strains carried AmpC genes, including blaDHA-1 found in K. pneumoniae (n=16) and E. cloacae (n=5) isolates, and blaEBC found in E. aerogenes (n=17) and E. coli (n=1) isolates, respectively. Eight out of 90 CRE isolates including 3 K. pneumoniae, 3 E. coli, 1 E. aerogenes and 1 E. cloacae without carbapenemase genes, but we detected ESBL and (or) AmpC genes in these strains, such as blaCTX-M-14, blaCTX-M-15, blaSHV-11, blaTEM-1 and blaDHA-1. As high as 41.1% (n=37) of the CRE isolates harbored carbapenemase, ESBL and together with AmpC genes, consisting of 17 E. aerogenes (56.7%), 14 K. pneumoniae (56.0%), 5 E. cloacae and 1 E. coli. However, no mcr gene was identified in this study.
In vitro Antimicrobial Susceptibility
All CRE isolates showed high resistance to the third or fourth-generation cephalosporins and carbapenemases (97.8%-100.0%) (Table 3), as well as cefoperazone-sulbactam (98.9%) and piperacillin–tazobactam (98.9%). The resistance rate of ciprofloxacin and trimethoprim-sulfamethoxazole was 93.3% and 53.3%, respectively. A small portion of Metallo-β-lactamases (MBL)-producing CRE isolates showed resistance to amikacin (15.7%) and gentamicin (31.4%), but the result was quite opposite among KPC-2 containing strains (83.3% and 91.7%) (p<0.05). In addition, 12.9% (9/70) of MBL-containing strains were found to be sensitive to aztreonam. Ceftazidime-avibactam showed better activity against the KPC-2-producing CRE isolates than the MBL-producing strains (p<0.05). The in vitro activity of fosfomycin and colistin and tigecycline against CRE isolates were 66.7%, 73.3% and 98.9%, respectively. The results are shown in Table 3. All CRE isolates were categorized into MDR in this study.
 |
Table 3 Antimicrobial Susceptibility and MIC Distributions of CRE Isolates |
Plasmid Replicon Types
For all CRE isolates, the IncX3 was the most common plasmid replicon type (76.7%, 69/90), and mainly found in NDM-containing strains (91.3%, 63/69) (Figure 2), followed by IncFII plasmid (37.8%, 34/90), and others including IncN (n=29), IncFIB (n=15), IncHI2 (n=2) and IncA/C (n=2). IncX3, IncN and IncFII were found together in 19 out of 56 NDM-5-producing isolates. As high as 91.7% (11/12) of KPC-2-producing strains simultaneously harbored the plasmids of IncFIB and IncFII, IncHI2 (n=2) was also detected. IncN was the unique plasmid found in the IMP-producing strains; however, IncX3, IncA/C and IncFIB plasmids were both detected in NDM-1 carrying strains. In addition, there were five strains without plasmid in this study.
 |
Figure 2 Dendrogram obtained from ERIC-PCR fingerprinting and Plasmid Replicon Profile of E. aerogenes (A), K. pneumoniae (B) and E. coli (C) isolates. |
STs and ERIC Patterns
As depicted in Figure 2, all NDM-5 producing E. aerogenes isolates belonged to ST4 and were classified into 12 different clusters (A1-12). Several strains among cluster A1, A4, A6, A9 and A11 had identical ERIC patterns and mainly distributed in NICU. Another E. aerogenes without carbapenemase gene were belonged to ST37. For K. pneumoniae isolates, ST11 (40%,10/25) was the dominant type and all found in KPC-2-producing strains, which was categorized into 4 clusters (B1-B4), with cluster B1 and B3 belonged to PICU (n=3) and gastroenterology ward (n=4), respectively, followed by ST48 which was detected in two NDM-5 producing K. pneumoniae isolates in gastroenterology ward and divided into cluster B10. Almost all strains in cluster B1, B3 and B10 were defined as the identical strains. Others including ST681, ST692, ST215, ST441, ST40, ST1428, ST690, ST17, ST1308, ST419, ST595 and ST2735. A total of seven STs were found among E. coli isolates, with ST692 (55.6%,10/18) being the prime type in NDM-5 containing strains and grouped into 3 different clusters (C10-C12), which were mainly prevalent in NICU (n=6), PICU (n=2) and gastroenterology ward (n=2), with many identical strains found as well. In addition, ST662 (n=3), ST2 (n=3), ST39 (n=2), ST35 (n=2), ST58 (n=2) and ST3 (n=1) were also detected. Meanwhile, diverse STs were identified between 9 E. Cloacae strains, including ST419, ST524, ST1318, ST920, ST391, ST45, ST39 and ST45.
Discussion
The retrospective epidemiological study was conducted for determining the prevalence and molecular epidemiology of inpatients with CRE intestinal colonization in a pediatric hospital of Shanghai, which helps us to figure out the current situation of fecal carriage of CRE among hospitalized children in china and take more scientific infection prevention and control measures. The fecal carriage rate of inpatients was 8.6% in our study, which was higher than two hospitals in Hunan (8.5%) and Fujian (6.6%), China.24,25 Worldwide, the colonization rate ranged from 18.9% to 69.5% in hospitalized patients.26–28 However, in outpatients, the rate of colonization with CRE in fecal sample collected from children was only 3.6% in our hospital in 2016.29 Consistent with the previous reports, CRE colonization in patients from community settings was infrequent. Healthcare facilities are usually considered as reservoirs of transmission30 of CRE, and the healthcare providers are partly to blame in CRE acquisition among hospitalized patients.31 Moreover, antibiotic exposures and invasive practices such as mechanical ventilation, percutaneous intervention and surgery increase the probability of CRE colonization and infection,32,33 and more than half CRE colonizers had such experiences before CRE being detected in this study. The CRE strains mainly distributed in NICU and PICU in our hospital, which might be related to critical underlying diseases, prolonged hospitalization, long-term application of carbapenems and other antimicrobial agents in these departments.34 In addition, five neonates were identified to carry CRE at birth, which may be attributed to the mother-to-infant transmission,35 and this remained to be further explored.
Enterobacteriaceae such as E. coli and K. pneumoniae are the part of the normal human intestinal flora but often responsible for HAI,36 and had become the predominant CRE isolates according to the Nationwide Surveillance data of Clinical CRE Strains in China,37,38 and were also detected in fecal samples in our study, mostly from patients in PICU and NICU. We should attach great importance to this, as ICU patients with these isolates were more likely to develop subsequent infection compared to patients without these isolates according to previous reports.39,40 It is noteworthy that E. aerogenes were the most dominant colonized CRE strains in children in this research. According to a recent study reported by Fupin Hu, etc., in 2020,41 the most prevalent CRE strains isolated from adult patients was K. pneumoniae (64.6%) in China from 2016 to 2018, and was consistent with the epidemic situation in our hospital before 2018 in clinic. While all 90 CRE strains were isolated from fecal samples of hospitalized children in 2019, and there was an outbreak of E. aerogenes between inpatient children (unpublished data). However, it still needs a further study on whether there is a causal relationship between clinical and colonized CRE strains, which would be explored in our following research.
Since the discovery of an ST14 K. pneumoniae with NDM-1 from a Swedish patient traveled to New Delhi in 2008,42 NDM-type MBLs have been sporadic occurrences in many regions all over the world (except the Middle East and Balkan countries).43 However, in china, blaNDM was the dominant carbapenemase gene found in children group clinically, and most prevalent in E. coli isolates.41 In our study, blaNDM-5 and blaNDM-1 genes were both identified, with blaNDM-5 being the main carbapenemase genotype, but detected more in E. aerogenes than in E. coli isolates. The blaKPC-2 was found the second most frequently and all found in K. pneumoniae isolates, which had spread across China and many other countries, causing both outbreaks and endemicity in certain regions.44 It is noteworthy that blaKPC-2 and blaNDM-5 had been the predominant carbapenemase genes detected in CR-KPN isolates from clinical specimens in our hospital in 2018.45 Besides, blaIMP-4 and blaIMP-26 were also observed, and 4 strains were identified to co-harboring blaNDM-1 and blaIMP-4, illustrating these genes spread in readily accessed between Enterobacteriaceae bacteria.
The mechanisms of carbapenem resistance in enterobacteriaceae include production of carbapenemases, β-lactamase (ESBLs and AmpC) activity combined with the mutation of porins, drug efflux pumps and alterations in penicillin-binding proteins. The first two are main mechanisms. In 8 CRE isolates without carbapenemase gene, we detected ESBLs and (or) AmpC genes, which might be one of the major causes of carbapenem resistance, others need to be further explored. All these resistance mechanisms of CRE strains are posing great challenges to clinical treatment.
All CRE isolates showed high resistance to cephalosporins and carbapenemases, as well as cefoperazone-sulbactam and piperacillin–tazobactam, which might be due to the widespread use of these antibacterial agents in children. As far as we all know, ceftazidime-avibactam has activity against serine β-lactamases but does not possess activity against MBL-producing organisms,46 therefore showing better activity against the KPC-2 producing CRE isolates than the MBL-producers (p<0.05). Fosfomycin, colistin and tigecycline, as the last-resort antibiotics in all to treat MDR infections,47 wherein tigecycline shows the highest (98.9%) activity in vitro. The increasing prevalence of CRE strains are attributed to the dissemination of conservative mobile elements carrying blaNDM or blaKPC-2 on plasmids in our country,37 consistent with the uptrend worldwide among NDM-containing isolates from clinical samples recently,45,48,49 IncX3 plasmid was the most common type in those strains. And our previous study revealed that the IncX3-type plasmid was responsible for the horizontal gene transfer of the blaNDM-5 gene among different Enterobacteriaceae isolates.50 IncFIB and IncFII were the predominant plasmids found in KPC-2-producing K. pneumoniae isolates and had reported to mediate clinical transmission in different districts of China.45,51 In our study, 100% of NDM-5 producing E. aerogenes and 55.6% of NDM-5 producing E. coli strains belonged to ST4 and ST692, respectively, and ST11 was the predominant clone-type in KPC-2 producing K. pneumoniae isolates. Several isolates of each species grouped into the same cluster were defined as the identical strains, indicating an ongoing nosocomial clonal transmission in our hospital in the same period, which may be acquired via physical contact with patients colonized or infected with CRE. Besides, two NDM-5 producing K. pneumoniae isolates belonged to ST48 also had the same ERIC pattern, and were reported to cause nosocomial outbreak in our hospital in 2017.52
CRE colonization is a prerequisite for development of CRE infection. According to a recent study conducted in Children’s Hospital of Fudan University,53 active CRE colonization surveillance and CRE positive patient propriety placement may decrease the CRE infection risk. Although the medical resources in many developing countries are limited, it is very important to take infection prevention and control measures to reduce the occurrence of nosocomial outbreak of CRE.
Conclusion
In conclusion, the study demonstrated a major intestinal colonization of ST4 NDM-5 E. aerogenes, ST11 KPC-2 K. pneumoniae and ST692 NDM-5 E. coli strains among hospitalized children in Shanghai, China. To reduce the nosocomial CRE infection and transmission, prevention and control measures should be strongly advocated and strictly implemented in clinical settings as soon as possible, especially in ICU.
Data Sharing Statement
All data generated and/or analyzed during the study are available from the corresponding author upon reasonable request.
Consent for Publication
All authors have consent for the manuscript publication.
Acknowledgments
We thank all members of the Clinical Laboratory of Shanghai Children’s Hospital for their cooperation and technical help.
Funding
This work was supported by the Shanghai Municipal Key Clinical Specialty (shslczdzk06902) and Shanghai Municipal Health Commission (201940253). The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–67. doi:10.1093/cid/cir202
2. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi:10.1016/S1473-3099(13)70190-7
3. Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi:10.1007/s10096-019-03673-1
4. Perez F, Villegas MV. The role of surveillance systems in confronting the global crisis of antibiotic-resistant bacteria. Curr Opin Infect Dis. 2015;28(4):375–383. doi:10.1097/QCO.0000000000000182
5. Feil EJ. Enterobacteriaceae: joining the dots with pan-European epidemiology. Lancet Infect Dis. 2017;17(2):118–119. doi:10.1016/S1473-3099(16)30333-4
6. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. doi:10.1016/j.molmed.2012.03.003
7. Goodman KE, Simner PJ, Tamma PD, Milstone AM. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther. 2016;14(1):95–108. doi:10.1586/14787210.2016.1106940
8. Kitchel B, Rasheed JK, Patel JB, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53(8):3365–3370. doi:10.1128/AAC.00126-09
9. Logan LK, Weinstein RA. the epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
10. McConville TH, Sullivan SB, Gomez-Simmonds A, Whittier S, Uhlemann AC. Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One. 2017;12(10):e0186195. doi:10.1371/journal.pone.0186195
11. ECDC. Carbapenem-resistant Enterobacteriaceae - first update;2018. Available from: https://ecdceuropaeu/en/publications-data/rapid-risk-assessment-carbapenem-resistant-enterobacteriaceae-first-update.
12. Joo EJ, Kim SJ, Baek M, et al. Fecal carriage of antimicrobial-resistant Enterobacteriaceae in Healthy Korean Adults. J Microbiol Biotechnol. 2018;28(7):1178–1184. doi:10.4014/jmb.1801.12060
13. Huang YS, Lai LC, Chen YA, et al. Colonization with multidrug-resistant organisms among healthy adults in the community setting: prevalence, risk factors, and composition of gut microbiome. Front Microbiol. 2020;11:1402. doi:10.3389/fmicb.2020.01402
14. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
15. Doosti A, Pourabbas M, Arshi A, Chehelgerdi M, Kabiri H. TEM and SHV genes in klebsiella pneumoniae isolated from cockroaches and their antimicrobial resistance pattern. Osong Public Health Res Perspect. 2015;6(1):3–8. doi:10.1016/j.phrp.2014.10.011
16. Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi:10.1128/JCM.40.6.2153-2162.2002
17. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
18. Hamouda A, Amyes SG. Development of highly ciprofloxacin-resistant laboratory mutants of Acinetobacter baumannii lacking topoisomerase IV gene mutations. J Antimicrob Chemother. 2006;57(1):155–156. doi:10.1093/jac/dki397
19. Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6):17–00672. doi:10.2807/1560-7917.ES.2018.23.6.17-00672
20. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi:10.1016/j.mimet.2005.03.018
21. Johnson TJ, Bielak EM, Fortini D, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant. Enterobacteriaceae Plasmid. 2012;68(1):43–50. doi:10.1016/j.plasmid.2012.03.001
22. Moosavian M, Emam N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect Drug Resist. 2019;12:1001–1010. doi:10.2147/IDR.S192597
23. Ranjbar R, Mirsaeed Ghazi F. Antibiotic sensitivity patterns and molecular typing of Shigella sonnei strains using ERIC-PCR. Iran J Public Health. 2013;42(10):1151–1157.
24. Liu Q, Liu L, Li Y, Chen X, Yan Q, Liu WE. Fecal carriage and epidemiology of carbapenem resistant enterobacteriaceae among hospitalized patients in a University Hospital. Infect Drug Resist. 2019;12:3935–3942. doi:10.2147/IDR.S233795
25. Zhao ZC, Xu XH, Liu MB, Wu J, Lin J, Li B. Fecal carriage of carbapenem-resistant Enterobacteriaceae in a Chinese university hospital. Am J Infect Control. 2014;42(5):e61–e64. doi:10.1016/j.ajic.2014.01.024
26. Prasad N, Labaze G, Kopacz J, et al. Asymptomatic rectal colonization with carbapenem-resistant Enterobacteriaceae and Clostridium difficile among residents of a long-term care facility in New York City. Am J Infect Control. 2016;44(5):525–532. doi:10.1016/j.ajic.2015.11.021
27. Singh K, Mangold KA, Wyant K, et al. Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J Clin Microbiol. 2012;50(8):2596–2600. doi:10.1128/JCM.00654-12
28. Papadimitriou-Olivgeris M, Christofidou M, Fligou F, et al. The role of colonization pressure in the dissemination of colistin or tigecycline resistant KPC-producing Klebsiella pneumoniae in critically ill patients. Infection. 2014;42(5):883–890. doi:10.1007/s15010-014-0653-x
29. Pan F, Tian D, Wang B, et al. Fecal carriage and molecular epidemiology of carbapenem-resistant Enterobacteriaceae from outpatient children in Shanghai. BMC Infect Dis. 2019;19(1):678. doi:10.1186/s12879-019-4298-3
30. Goodman KE, Simner PJ, Klein EY, et al. How frequently are hospitalized patients colonized with carbapenem-resistant Enterobacteriaceae (CRE) already on contact precautions for other indications? Infect Control Hosp Epidemiol. 2018;39(12):1491–1493. doi:10.1017/ice.2018.236
31. Grabowski ME, Kang H, Wells KM, Sifri CD, Mathers AJ, Lobo JM. Provider role in transmission of carbapenem-resistant Enterobacteriaceae. Infect Control Hosp Epidemiol. 2017;38(11):1329–1334. doi:10.1017/ice.2017.216
32. Bogan C, Kaye KS, Chopra T, et al. Outcomes of carbapenem-resistant Enterobacteriaceae isolation: matched analysis. Am J Infect Control. 2014;42(6):612–620. doi:10.1016/j.ajic.2014.02.013
33. Chiotos K, Tamma PD, Flett KB, et al. Multicenter study of the risk factors for colonization or infection with carbapenem-resistant Enterobacteriaceae in children. Antimicrob Agents Chemother. 2017;61(12):e01440–17. doi:10.1128/AAC.01440-17
34. Salomão MC, Freire MP, Boszczowski I, Raymundo SF, Guedes AR, Levin AS. Increased risk for carbapenem-resistant enterobacteriaceae colonization in intensive care units after hospitalization in emergency department. Emerg Infect Dis. 2020;26(6):1156–1163. doi:10.3201/eid2606.190965
35. Principe L, Meroni E, Conte V, et al. Mother-to-child transmission of KPC-producing Klebsiella pneumoniae: potential relevance of a low microbial urinary load for screening purposes. J Hosp Infect. 2018;98(3):314–316. doi:10.1016/j.jhin.2017.10.008
36. van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. 2016;30(2):377–390. doi:10.1016/j.idc.2016.02.004
37. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
38. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62(2):e01882–17.
39. Freedberg DE, Zhou MJ, Cohen ME, et al. Pathogen colonization of the gastrointestinal microbiome at intensive care unit admission and risk for subsequent death or infection. Intensive Care Med. 2018;44(8):1203–1211. doi:10.1007/s00134-018-5268-8
40. Gorrie CL, Mirceta M, Wick RR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65(2):208–215. doi:10.1093/cid/cix270
41. Han R, Shi Q, Wu S, et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among Carbapenem-Resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
42. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla (NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
43. Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int. 2014;2014:249856. doi:10.1155/2014/249856
44. Chitnis AS, Caruthers PS, Rao AK, et al. Outbreak of carbapenem-resistant enterobacteriaceae at a long-term acute care hospital: sustained reductions in transmission through active surveillance and targeted interventions. Infect Control Hosp Epidemiol. 2012;33(10):984–992. doi:10.1086/667738
45. Wang B, Pan F, Wang C, et al. Molecular epidemiology of Carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int J Infect Dis. 2020;93:311–319. doi:10.1016/j.ijid.2020.02.009
46. Zasowski EJ, Rybak JM, Rybak MJ. The β-Lactams strike back: ceftazidime-Avibactam. Pharmacotherapy. 2015;35(8):755–770. doi:10.1002/phar.1622
47. Davido B, Bouchand F, Dinh A, et al. Reinforcement of an antimicrobial stewardship task force aims at a better use of antibiotics of last resort: the COLITIFOS study. Int J Antimicrob Agents. 2017;50(2):142–147. doi:10.1016/j.ijantimicag.2017.03.030
48. Paskova V, Medvecky M, Skalova A, et al. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech Hospitals. Front Microbiol. 2018;9:1549. doi:10.3389/fmicb.2018.01549
49. Xu J, He F. Characterization of an NDM-7 carbapenemase-producing Escherichia coli ST410 clinical strain isolated from a urinary tract infection in China. Infect Drug Resist. 2019;12:1555–1564. doi:10.2147/IDR.S206211
50. Tian D, Wang B, Zhang H, et al. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among enterobacteriaceae in children. mSphere. 2020;5(1):e00699–19. doi:10.1128/mSphere.00699-19
51. Zhou H, Zhang K, Chen W, et al. Epidemiological characteristics of carbapenem-resistant Enterobacteriaceae collected from 17 hospitals in Nanjing district of China. Antimicrob Resist Infect Control. 2020;9:15. doi:10.1186/s13756-019-0674-4
52. Tian D, Pan F, Wang C, Sun Y, Zhang H. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai [published correction appears in Infect Drug Resist]. Infect Drug Resist. 2018;11:1935–1943. doi:10.2147/IDR.S175584
53. Yin L, He L, Miao J, et al. Actively surveillance and appropriate patients placements’ contact isolation dramatically decreased Carbapenem-Resistant Enterobacteriaceae infection and colonization in pediatric patients in China. J Hosp Infect. 2020;S0195–6701(20):30130–30134.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
