Back to Journals » OncoTargets and Therapy » Volume 14
Favorable Response to Olaparib in a Patient with Cancer of Unknown Primary Carrying a Germline BRCA1 R71K Mutation
Authors Jia X, Zhao S, Li X, Lv L, Chen X , Pan E, Ou Q , Song C, Sun S, Zhao J, Xu L, Li M
Received 18 August 2021
Accepted for publication 16 November 2021
Published 27 November 2021 Volume 2021:14 Pages 5353—5360
DOI https://doi.org/10.2147/OTT.S334847
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Xiaomeng Jia,1,* Shanshan Zhao,1,* Xiang Li,2,* Li Lv,3,* Xin Chen,4 Evenki Pan,4 Qiuxiang Ou,4 Chen Song,1 Siwen Sun,1 Jinbo Zhao,1 Lingzhi Xu,1 Man Li1
1Department of Oncology, The Second Hospital of Dalian Medical University, Dalian, 116023, People’s Republic of China; 2Department of Radiology, The Second Hospital of Dalian Medical University, Dalian, 116023, People’s Republic of China; 3Department of Pathology, The Second Hospital of Dalian Medical University, Dalian, 116023, People’s Republic of China; 4Geneseeq Research Institute, Nanjing Geneseeq Technology Inc., Nanjing, 210032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Man Li; Lingzhi Xu
Department of Oncology, The Second Hospital of Dalian Medical University, Dalian, 116023, People’s Republic of China
Tel +86-17709873580
; +86-17709873532
Email [email protected]; [email protected]
Abstract: The treatment options for cancer of unknown primary (CUP) are challenging due to the lack of knowledge about the primary sites, often resulting in a poor prognosis. The emerging next-generation sequencing (NGS) technique has provided a reliable approach to facilitate tumor primary site prediction and targetable gene alteration identification for CUP patients. In this report, we described a 63-year-old female patient who experienced recurrent CUP. NGS-based genetic profiling results revealed a pathogenic germline BRCA1 R71K mutation. Accordingly, the patient received the poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitor olaparib treatment and demonstrated a favorable response to this treatment. Our case suggests that NGS holds great promise for providing improved diagnosis and treatment options to patients with CUP, warranting further clinical investigation.
Keywords: cancer of unknown primary, BRCA1 germline mutation, olaparib, next-generation sequencing
Introduction
Cancer of unknown primary (CUP) is a malignant metastatic disease, the primary sites for which cannot be identified at the time of diagnosis despite rigorous efforts.1 Although the development of detection technologies could improve the identification of the primary site, CUP remains the sixth to eighth most common cancer globally, accounting for 2–5% of all diagnosed malignancies.1,2 Epidemiological studies have shown that the median age of diagnosed CUP patients is 60 with no significant sex difference, and over 80% of patients diagnosed with CUP displayed an aggressive form of the disease.3 The absence of a primary tumor for CUP can be a result of its size, dormancy, and involution, and such cryptic pathogenesis imposes challenges on the treatment of CUP.2 Due to its late onset, difficulty in diagnosis, limited treatment options, and strong drug resistance, the prognosis of CUP is often depressing with a median survival of less than one year.1 A study found patients with CUP experienced significantly poorer survival than those with metastatic cancer of a known primary site.4 Indeed, CUP is the third to fourth most common cause of cancer-related mortality.1
Germline BReast CAncer 1/2 (BRCA1/2) mutations place patients at a high risk of developing breast cancer, ovarian cancer, prostate cancer, melanoma, and pancreatic cancer with clinicopathologic features of early onset, high grade, and aggressiveness.5,6 The BRCA genes are essential for the homologous recombination (HR) pathway, which functions in high-fidelity DNA double-strand break (DSB) repair.7 HR deficiency (HDR) due to inactivation of the BRCA genes can result in genome instability, and germline BRCA1/2 mutation status is currently one of the most relevant genetic tumor markers for HRD used in the clinic.7–9 As sensitivity to platinum drugs is a feature of HRD, the HRD patients are likely to benefit from the poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibition by the strategy of synthetic lethality.10 In detail, tumors with defects in BRCA1/2 have compromised ability to repair DNA DSB by HR, and they are then highly sensitive to blockade of the DNA single-strand break repair caused by the inhibition of PARP enzymatic activity.11,12 Over the past decade, four PARP inhibitors, including olaparib, rucaparib, niraparib and talazoparib, have been approved by the FDA for clinical use as single agents.13
Currently, next-generation sequencing (NGS) has facilitated the prediction of tumor primary sites and the identification of targetable gene mutations in patients with CUP. The recent clinical trial has shown the promise of site-specific treatment and targeted therapy based on NGS testing results for CUPs, which warrants further clinical investigation.14 Herein, we report that a 63-year-old female patient who experienced recurrent CUP showed durable clinical benefit from the olaparib treatment in light of the pathogenic germline BRCA1 mutation as identified by the NGS genetic testing.
Case Report
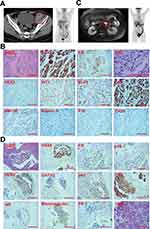
A 63-year-old, female new patient was admitted to the hospital in October 2012, and the timeline of her clinical records and treatments throughout the 9-year course are demonstrated in Figure 1. The patient presented with an egg-sized lump in her left iliac fossa (Figure 2A, left). The patient has no family history of cancer except that her mother died of a malignant tumor of unknown primary. Laboratory examinations revealed that her pretreatment carbohydrate antigen CA125 level was 133.6 U/mL, whereas no abnormalities were observed with the levels of carcinoembryonic antigen (CEA), carbohydrate antigen CA19-9, alpha-fetoprotein (AFP), and carbohydrate antigen CA72-4. The patient underwent resection of the lump, and the pathological results (Figure 2B) are as follows: Haemotoxylin and eosin (H&E) staining revealed the histologic pattern of adenocarcinoma. Immunohistochemical (IHC) staining showed tumor proliferation marker antigen KI-67 (Ki-67, 70%), tumor marker p53 (diffuse +), squamous carcinoma marker p40 (sporadic +), intestinal adenocarcinoma marker cytokeratin 20 (CK20, -), ovarian cancer marker Wilms tumor 1 (WT1, +), high-grade cervical squamous intraepithelial lesion marker p16 (sporadic +), ovarian clear cell carcinoma marker, hepatocyte nuclear factor-1-beta (HNF1B, -) and Napsin A (-), and the Paired Box 8 (PAX8, +) marker for renal, Müllerian, and thyroid carcinomas. The site of the primary tumor was unclear based on the pathological evidence. Subsequent positron emission tomography-computed tomography (PET-CT) examinations (Figure 2A, right) revealed increased fluorodeoxyglucose (FDG) metabolism in the right sterno-diaphragmatic angle and cardiophrenic angle, right crus of the diaphragm, retroperitoneum, and left pelvic cavity, which was initially considered as metastatic tumors. The slight increase in FDG metabolism at the lower left abdominal wall and the small lymph nodes of the left groin was first considered as postoperative changes. The increased FDG metabolism in the third and fourth posterior lumbar spine was considered as the inflammatory response. A liver cyst was detected. Further pathological examination was warranted for the small convex perineal nodules with increased FDG metabolism. In November 2012, the patient received the paclitaxel–cisplatin chemotherapy regimen for a total of 6 cycles. During the treatment, the tumor marker CA125 gradually decreased to the normal range (0–35 U/mL), and the therapeutic efficacy was evaluated as partial response (PR). After chemotherapy, the patient was monitored by regular imaging examinations with no sign of tumor recurrence or metastasis, and her CA125 level remained in the normal range.
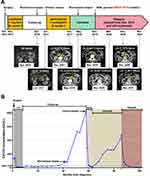 |
Figure 1 Timeline of treatment with radiography of responses and CA125 concentration. (A) Timeline of treatment and corresponding MRI. (B) Timeline of treatment and corresponding CA125 concentration. |
In April 2016, her CA125 level was found to reach 84.7 U/mL. Given no clear sign of tumor recurrence or metastasis according to imaging examinations, we considered this case as biochemical recurrence and did not pursue the remaining therapy. In July 2017, the CA125 level increased to 824.2 U/mL, and magnetic resonance imaging (MRI) indicated enlarged lymph nodes in the retroperitoneal area (Figure 2C, left). The patient then received gemcitabine-carboplatin regimen chemotherapy for a total of 6 cycles, and the CA125 level decreased to the normal range again. CT examination showed the size of retroperitoneal lymph nodes significantly reduced after chemotherapy (December 2017), and the efficacy was evaluated as PR. After 6 cycles of chemotherapy, we revisited the tumor specimen from her left iliac fossa, added additional IHC tests, and found estrogen receptor (ER), progesterone receptor (PR), and Human Epidermal Growth Factor Receptor 2 (HER2) were all positive (70% medium +, 5% weak +, and 1+, respectively). The positivity suggested that the primary tumor site could be located in female genital organs or the breast. Due to her poor tolerance to chemotherapy, the patient started maintenance therapy with oral letrozole.
In May 2018, the patient’s CA125 level elevated to 90.9 U/mL. CT results showed the retroperitoneal lymph nodes resembled her biochemical recurrence scenario in December 2017. The patient continued letrozole treatment until CA125 reached 481.6 U/mL in May 2019. CT results revealed significantly enlarged retroperitoneal lymph nodes compared to May 2018, so the efficacy was evaluated as progressive disease (PD). In May 2019, the patient underwent the PET-CT examination (Figure 2C, right), which revealed multiple retroperitoneal lymph nodes with high FDG metabolism, suggestive of metastasis. Due to inflammation in the colon, the patient underwent the colonoscopy examination and began to take the ulcerative colitis treatment accordingly. In November 2019, she received the CT-guided retroperitoneal lymph node puncture operation, and the pathological results of the lymph node biopsy are as follows (Figure 2D). H&E staining indicated poorly differentiated adenocarcinoma, and some of the cancer cells had clear cytoplasm. IHC staining showed p40 (-), PAX8 (diffuse strong +), squamous carcinoma marker p63 (-), intestinal adenocarcinoma marker cytokeratin 7 (CK7, +), and breast carcinoma marker GATA Binding Protein 3 (GATA3, a little cytoplasmic +). Together with the markers of ER (40%+), PR (-), HER2 (2+), androgen receptors (AR, ~5%+), and mammaglobin (-), it was likely to be adenocarcinoma of the female genital organs. However, the primary site was still undetermined.
The patient underwent targeted NGS using the 48 cancer gene MAMECAN™ panel in a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited clinical testing laboratory (Nanjing Geneseeq Technology, Nanjing, China) since November 2019 (Table S1). Targeted NGS was performed as previously described.15 In brief, germline mutations were identified from genomic DNA extracted from whole blood control samples using the DNeasy Blood & Tissue Kit (Qiagen). Cell-free DNA (cfDNA) was extracted from the plasma supernatant using QIAamp Circulating Nucleic Acid Kit (Qiagen) for circulating tumor DNA (ctDNA) analysis. NGS library preparation was performed using KAPA Hyper Prep Kit (KAPA Biosystems, USA). Customized xGen lockdown probes (Integrated DNA Technologies) targeting the cancer-relevant genes were used for hybridization capture enrichment. Enriched libraries were on-beads PCR amplified, purified, and sequenced on the Illumina HiSeq4000 platform. The NGS test revealed a pathogenic, heterozygous germline BRCA1 331G>A nucleotide substitution mutation, which results in an Arg-to-Lys change (Figure 3A). Therefore, the patient began oral olaparib treatment at 300 mg twice a day from December 2019. The patient has demonstrated PR to olaparib, and the CA125 level dropped from 228.8 U/mL to 6.0 U/mL after the treatment. The following CT examinations in March 2020, November 2020, and March 2021 all confirmed progressive shrinkage of retroperitoneal lymph nodes (Figure 1A). Molecularly, we detected one ClinVar database-defined “likely pathogenic” somatic mutation S241C in tumor suppressor TP53 at a mutant allele frequency of 2.97% from the November 2019 NGS test (Table S2). Another somatic mutation CHEK2 G178E was also detected by NGS, but its biological significance was unknown. With olaparib treatment, we observed effective elimination of ctDNA as the TP53 S241C mutation was no longer detected in the two subsequent NGS tests (Figure 3B). Up till present, the patient has been on the olaparib monotherapy for 15 months and remained progression-free with good tolerability and high quality of life.
 |
Figure 3 Diagrams of mutations identified in NGS testing. (A) Schematic diagram of the R71K mutation in full-length BRCA1 (adopted from cBioPortal MutationMapper https://www.cbioportal.org/mutation_mapper, retrieved on Nov. 3, 2021). (B) Abundance change of TP53 S241C in the time course of olaparib treatment. The ctDNA abundance measured by each NGS test is represented by the dot. |
Discussion
Accurate determination of the tissue of origin for metastatic tumors is important for effective treatment and improved patient outcome, but it often remains elusive for CUPs, which imposes a therapeutic challenge.16 In the era of precision medicine, assays such as imaging and immunohistochemical evaluation and molecular profiling, especially emerging genomic profiling, have been utilized to predict the primary site and highlighted the advantage of personalized therapy.16 For instance, gene expression and alterations from NGS may help predict the primary site of CUPs and guide treatment, and such targeted therapy for CUP patients has been in the clinical trial.14 A number of case reports have confirmed the significance of NGS for CUPs. Kato et al reported a case of unknown primary adenocarcinoma with KRAS G12D and MLH1 R389W mutations identified by NGS-based ctDNA evaluation, and the clinical efficacy reached PR after targeted application of Trametinib and Nivolumab.17 Subbiah et al reported 7 cases of CUP all benefited from NGS-guided, targeted therapy.18 Both Ross and Zhao et al reported cases of CUP with EML4-ALK fusion found by NGS, and patients achieved PR after targeted therapy of the ALK inhibitor crizotinib.19,20 Mitani et al reported successful treatment of a CUP patient with EGFR mutations under the guidance of NGS, achieving long-term disease control.21
Here, we report a unique case of a CUP patient carrying a germline BRCA1 pathogenic mutation R71K, whereas the application of the PARP inhibitor olaparib has achieved good therapeutic effects. Although we performed extensive pathological examinations (eg, H&E stain and immunohistochemistry) and imaging technologies (eg, CT, MRI, and PET-CT scan), the primary site has not yet been identified, while additional workup such as direct internal organ visualization by an exploratory laparotomy may facilitate a definitive diagnosis.22 The use of empirical first- and second-line chemotherapies initially reached acceptable results, but the patient later suffered from poor tolerance and disease recurrence. Endocrine therapy with letrozole based on IHC results exhibited limited efficacy. Subsequently, the NGS-based profiling detected the BRCA1 R71K point mutation. Olaparib was administered accordingly, and the efficacy has been favorable as evidenced by reduction of CA125 to a normal level, shrinkage of the retroperitoneal lymph nodes, and elimination of detected plasma ctDNA. The patient has been evaluated as PR during long-term follow-up till now.
BRCA1/2 are tumor suppressor genes maintaining genome integrity by regulating homologous recombination repair (HRR).23 Germline pathogenic variants in BRCA genes are associated with a higher risk of cancer in an autosomal dominant manner.23 Although the prognostic value of BRCA1/2 mutations for cancer patients remains controversial, ovarian cancer patients carrying inherited BRCA gene mutations were found to have better survival compared to non-carriers, regardless of tumor stage, grade, or histologic subtype.24 A large-scale molecular profiling across 21 tumor lineages composed of 52,426 tumor samples revealed that BRCA1/2 was among the top commonly mutated HR-related genes harboring pathogenic mutations.5 Recent studies suggested that BRCA mutations have a prevalence of 2–8% among breast cancer patients, 13–15% among ovarian cancer patients, and a population carrier frequency of 0.2–1%.25 The types of BRCA disease-causing mutations include frameshift, stop codon, rearrangement, missense, and splice variant mutations.26
The BRCA1 331G>A (GenBank: U14680.1) point mutation we reported in the exon 5 of BRCA1 can theoretically result in an Arg-to-Lys change at codon 71 (R71K).27 Previous research has shown that this mutation significantly increased the BRCA1 transcript containing a 22-bp deletion in exon 5 and putatively encoded a truncated BRCA1 protein of 63 amino acids instead of 1863 residues.27 It is likely because the 331G>A mutation is located at the last nucleotide of exon 5 and alters the guanidine of the 5ʹ splice site consensus sequence “AG” leading to the use of an alternative splice site. This is a loss-of-function mutation that was also reported in other studies, resulting in disruption of the highly conserved zinc finger ring domain (residue 24–78) and loss of the downstream functional domains.27–30 These findings justified the treatment rationale for the patient receiving the PARP inhibitor olaparib and achieving a sustained clinical benefit.
Olaparib, the first PARP inhibitor approved by the FDA, acts as a competitive inhibitor of nicotinamide adenine dinucleotide (NAD+) at the catalytic sites of PARP1 and PARP2, leading to DNA damage and synthetic lethality with BRCA deficiency.31 Based on the results of SOLO-1 and SOLO-2 studies, olaparib was approved by the FDA for platinum-sensitive recurrent ovarian cancer.32,33 The OlympiAD’s results led to its approval for use in breast cancer.34 Since the trial POLO announced its results, the FDA has approved olaparib for pancreatic cancer patients with an inherited BRCA mutation.35 Based on the results of the trial PROfound, the FDA approved its use in metastatic castration-resistant prostate cancer (mCRPC) patients with HRR gene mutation.36 To date, applications of olaparib for a wider range of cancers are still under investigation and have shown great potential.
Conclusion
In conclusion, our case has demonstrated a favorable response to the olaparib treatment in a CUP patient carrying a germline BRCA1 R71K mutation. It underscores the application of NGS in guiding precision therapy and targeted treatment for CUPs with sophisticated histopathologic characteristics. Currently, most of such studies, including this one, are case reports based on the genomic status of individual patients, while the clinical value of the genomic changes in the CUP population needs to be systematically evaluated. Going forward, the clinical trials, including CUPISCO, OCTANE, and GENIUS,37–39 hold great promise to establish the role of NGS testing in the evaluation and treatment of patients with CUPs.
Data Sharing Statement
Additional data related to the genetic tests, pathologic reports, treatment information, and images are available for review upon reasonable request from the corresponding author.
Ethics Approval and Consent to Participate
Institutional approval was not required for publication of this case report. The authors hold written informed consent provided by the patient for publication of this case report. We would like to thank the patient for providing consent for publication. We also thank all staff involved in this case study.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (No. 81872156 to Man Li), Provincial Natural Science Foundation of Liaoning (No. 2019-MS-096 to Lingzhi Xu), and Technological Innovation Foundation of Dalian (No. 2019J13SN87 to Lingzhi Xu).
Disclosure
Xin Chen, Evenki Pan, and Qiuxiang Ou are employees of Nanjing Geneseeq Technology Inc., China. The remaining authors have no competing interests to declare.
References
1. Qaseem A, Usman N, Jayaraj JS, Janapala RN, Kashif T. Cancer of unknown primary: a review on clinical guidelines in the development and targeted management of patients with the unknown primary site. Cureus. 2019;11(9):e5552. doi:10.7759/cureus.5552
2. Hemminki K, Bevier M, Sundquist J, Hemminki A. Cancer of unknown primary (CUP): does cause of death and family history implicate hidden phenotypically changed primaries? Ann Oncol. 2012;23(10):2720–2724. doi:10.1093/annonc/mds063
3. Alshareeda AT, Al-Sowayan BS, Alkharji RR, Aldosari SM, Al Subayyil AM, Alghuwainem A. Cancer of unknown primary site: real entity or misdiagnosed disease? J Cancer. 2020;11(13):3919–3931. doi:10.7150/jca.42880
4. Kim CS, Hannouf MB, Sarma S, et al. Survival outcome differences based on treatments used and knowledge of the primary tumour site for patients with cancer of unknown and known primary in Ontario. Curr Oncol. 2018;25(5):307–316. doi:10.3747/co.25.4003
5. Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018;2:1–13. doi:10.1200/PO.17.00286
6. Ye F, He M, Huang L, et al. Insights into the impacts of BRCA mutations on clinicopathology and management of early-onset triple-negative breast cancer. Front Oncol. 2020;10:574813. doi:10.3389/fonc.2020.574813
7. Nguyen L, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11(1):5584. doi:10.1038/s41467-020-19406-4
8. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi:10.1038/nrc.2015.21
9. Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110(7):704–713. doi:10.1093/jnci/djy085
10. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. doi:10.1158/2159-8290.CD-15-0714
11. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi:10.1038/nature03445
12. Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi:10.1056/NEJMoa0900212
13. Chan CY, Tan KV, Cornelissen B. PARP inhibitors in cancer diagnosis and therapy. Clin Cancer Res. 2021;27(6):1585–1594. doi:10.1158/1078-0432.CCR-20-2766
14. Hayashi H, Takiguchi Y, Minami H, et al. Site-specific and targeted therapy based on molecular profiling by next-generation sequencing for cancer of unknown primary site: a nonrandomized phase 2 clinical trial. JAMA Oncol. 2020;6(12):1931–1938. doi:10.1001/jamaoncol.2020.4643
15. Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24(13):3097–3107. doi:10.1158/1078-0432.CCR-17-2310
16. Laprovitera N, Riefolo M, Ambrosini E, Klec C, Pichler M, Ferracin M. Cancer of unknown primary: challenges and progress in clinical management. Cancers. 2021;13(3):451. doi:10.3390/cancers13030451
17. Kato S, Krishnamurthy N, Banks KC, et al. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. 2017;77(16):4238–4246. doi:10.1158/0008-5472.CAN-17-0628
18. Subbiah IM, Tsimberidou A, Subbiah V, Janku F, Roy-Chowdhuri S, Hong DS. Next generation sequencing of carcinoma of unknown primary reveals novel combinatorial strategies in a heterogeneous mutational landscape. Oncoscience. 2017;4(5–6):47–56. doi:10.18632/oncoscience.352
19. Ross JS, Wang K, Gay L, et al. Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies. JAMA Oncol. 2015;1(1):40–49. doi:10.1001/jamaoncol.2014.216
20. Zhao P, Peng L, Wu W, et al. Carcinoma of unknown primary with EML4-ALK fusion response to ALK inhibitors. Oncologist. 2019;24(4):449–454. doi:10.1634/theoncologist.2018-0439
21. Mitani Y, Kanai M, Kou T, et al. Cancer of unknown primary with EGFR mutation successfully treated with targeted therapy directed by clinical next-generation sequencing: a case report. BMC Cancer. 2020;20(1):1177. doi:10.1186/s12885-020-07640-4
22. Sugarbaker PH. Peritoneal carcinomatosis of unknown primary site, a study of 25 patients over 30 years. Eur J Surg Oncol. 2020;46(10 Pt A):1908–1911. doi:10.1016/j.ejso.2020.03.217
23. Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. Adam MP, Ardinger HH, Pagon RA, et al. editors. Genereviews(R). Seattle: University of Washington; 1998. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1247/.
24. Zhong Q, Peng HL, Zhao X, Zhang L, Hwang WT. Effects of BRCA1- and BRCA2-related mutations on ovarian and breast cancer survival: a meta-analysis. Clin Cancer Res. 2015;21(1):211–220. doi:10.1158/1078-0432.CCR-14-1816
25. Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: distribution and clinical implications. Curr Opin Obstet Gynecol. 2010;22(1):72–78. doi:10.1097/GCO.0b013e328332dca3
26. Heramb C, Wangensteen T, Grindedal EM, et al. BRCA1 and BRCA2 mutation spectrum – an update on mutation distribution in a large cancer genetics clinic in Norway. Hered Cancer Clin Pract. 2018;16:3. doi:10.1186/s13053-017-0085-6
27. Zhang L, Chen L, Bacares R, et al. BRCA1 R71K missense mutation contributes to cancer predisposition by increasing alternative transcript levels. Breast Cancer Res Treat. 2011;130(3):1051–1056. doi:10.1007/s10549-011-1732-7
28. Meindl A; German Consortium for Hereditary Breast and Ovarian Cancer. Comprehensive analysis of 989 patients with breast or ovarian cancer provides BRCA1 and BRCA2 mutation profiles and frequencies for the German population. Int J Cancer. 2002;97(4):472–480.
29. Clark SL, Rodriguez AM, Snyder RR, Hankins GD, Boehning D. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J. 2012;1(1):e201204005. doi:10.5936/csbj.201204005
30. Fang M, Zhu L, Li H, et al. Characterization of mutations in BRCA1/2 and the relationship with clinic-pathological features of breast cancer in a hereditarily high-risk sample of Chinese population. Oncol Lett. 2018;15(3):3068–3074. doi:10.3892/ol.2017.7717
31. Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72(21):5588–5599. doi:10.1158/0008-5472.CAN-12-2753
32. Olaparib maintenance monotherapy in patients with BRCA mutated ovarian cancer following first line platinum based chemotherapy. Available from: https://ClinicalTrials.gov/show/NCT01844986.
33. Olaparib after response to trabectedin-pegylated liposomal doxorubicin in recurrent ovarian carcinoma. Available from: https://ClinicalTrials.gov/show/NCT03470805.
34. Assessment of the efficacy and safety of olaparib monotherapy versus physicians choice chemotherapy in the treatment of metastatic breast cancer patients with germline BRCA1/2 mutations. Available from: https://ClinicalTrials.gov/show/NCT02000622.
35. Olaparib in gBRCA mutated pancreatic cancer whose disease has not progressed on first line platinum-based chemotherapy. Available from: https://ClinicalTrials.gov/show/NCT02184195.
36. Study of olaparib (Lynparza™) versus enzalutamide or abiraterone acetate in men with metastatic castration-resistant prostate cancer (PROfound Study). Available from: https://ClinicalTrials.gov/show/NCT02987543.
37. A phase II randomized study comparing the efficacy and safety of targeted therapy or cancer immunotherapy versus platinum-based chemotherapy in patients with cancer of unknown primary site. Available from: https://ClinicalTrials.gov/show/NCT03498521.
38. Ontario-wide cancer targeted nucleic acid evaluation. https://ClinicalTrials.gov/show/NCT02906943. Available from:
39. Genomic investigation of unusual responders. Available from: https://ClinicalTrials.gov/show/NCT03740503.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.