Back to Journals » OncoTargets and Therapy » Volume 14
FAST1 Predicts Poor Survival of Renal Carcinoma and Promotes Its Progression Through the TGF-β/Smad Pathway
Authors Tian T, Fu X, Hu L, Yang X, Sun P, Sun F
Received 26 October 2020
Accepted for publication 18 January 2021
Published 26 February 2021 Volume 2021:14 Pages 1487—1499
DOI https://doi.org/10.2147/OTT.S288847
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Tao Tian,1 Xiangyang Fu,2 Liangliang Hu,1 Xiaofeng Yang,1 Peng Sun,1 Fengfeng Sun1
1Department of Urology, Zaozhuang Municipal Hospital, Zaozhuang, 277100, Shandong, People’s Republic of China; 2Zaozhuang Yicheng District People’s Hospital, Zaozhuang, Shandong, 277300, People’s Republic of China
Correspondence: Fengfeng Sun
Department of Urology, Zaozhuang Municipal Hospital, No. 41 Longtou Road, Shizhong District, Zaozhuang, 277100, Shandong, People’s Republic of China
Tel + 86 0632 3288037
Email [email protected]
Purpose: Renal carcinoma (RC) originates in the renal tubular epithelial system, among which renal cell carcinoma (RCC) is the most frequent one. The forkhead activin signal transducer 1 (FAST1) has been shown to interfere with tumor progression as an oncogene, while its role in RC is limited. Therefore, this paper explored the prognostic significance, specific effects, and related mechanisms of FAST1 on RC.
Patients and Methods: Cell colony formation assay, cell counting kit-8 (CCK8) assay, flow cytometry and Transwell assay were used to test cell proliferation, viability, apoptosis, migration and invasion, respectively. Western blot (WB) was employed to determine the protein level of FAST1.
Results: Our study confirmed that FAST1 was up-regulated in RC tissues and cell lines, and its overexpression often represented a poor prognosis of RC patients. Meanwhile, the in vitro experiments showed that overexpressing FAST1 facilitated RC cell viability, proliferation, migration, invasion and epithelial-mesenchymal transition (EMT), and repressed cell apoptosis. In addition, the in vivo experiments illustrated that the up-regulation of FAST1 strengthened tumor growth. On the contrary, knocking down FAST1 had the opposite effects. Mechanistically, The TGF-β/Smad pathway contributed to RC evolvement and was activated by FAST1 both in vitro and in vivo.
Conclusion: This article suggests that FAST1 exerts a carcinogenic role in RC by regulating the TGF-β/Smad signaling.
Keywords: forkhead activin signal transducer 1, TGF-β/Smad pathway, renal carcinoma
Introduction
Renal carcinoma (RC) is a highly malignant tumor of the urinary system, with insidious onset and high mortality. Renal cell carcinoma (RCC) is the most typical type of RC. Due to the lack of specific markers and typical clinical manifestations, early RC is not easily detected. Studies have revealed that the five-year survival rate of early RC patients is about 90%, while that of advanced RC patients falls to 10%.1 Worse still, advanced RC patients are not suitable for surgery and are resistant to chemoradiotherapy, with high metastasis and recurrence rate. Meanwhile, they are not sensitive to immunotherapy.2 Therefore, it is crucial to find new biomarkers of RC. This paper aims to study new RC markers to improve the poor prognosis of RC.
Forkhead activin signal transducer 1 (FOXH1), also called FAST1, is encoded by a gene located on 8q24.3. Functionally, FOXH1 acts as a primitive-streak specifier and activin co-effector, thus having a vital role in the development by promoting the generation of human-induced pluripotent stem cells (iPSCs) from somatic cells via cellular signaling transduction.3 Moreover, FOXH1 binds SMAD2 and activates an activin response element by binding the DNA motif TGT(G/T) (T/G) ATT.4 Recently, FAST1 has been found dysregulated in several tumors and contributes to tumor growth and metastasis. Some studies have demonstrated that the FAST1 level is positively related to the poor prognosis of colorectal cancer (CRC). Overexpressing FAST1 enhances cell proliferation, migration, invasion, and pulmonary metastasis of CRC.5 Additionally, Let-7b and let-7f inhibit cell proliferation by blocking the cell cycle of the G1-S transition through directly targeting FOXH1.6 However, the specific effects of FAST1 on RC, especially the interaction between FAST1 and different downstream pathways on RC, need to be further studied.
Transforming growth factor β (TGF-β) is a disulfide-bonded dimer protein containing three ligands, namely TGF-β1, TGF-β2 and TGF-β3. Current studies have found that TGF-β has a dual role in tumors. It inhibits tumors in the early stage while acts as a carcinogen in the advanced stage.7 Oshimori et al have illustrated that TGF-β plays a carcinogenic role in squamous cell carcinoma and that tumor cells that are highly sensitive to TGF-β have a higher invasion and metastasis rate and stronger drug resistance than those are less sensitive to TGF-β.8 Solvated metal atom dispersed (Smad) is one of the classic TGF-β dependent proteins with a length of about 500 amino acids, containing spherical MH1 and MH2 domains and a cohesive variable zone. Smad2, a vital member of the Smad family, often acts as an activator of TGF-β.9 Some scholars have found that the activated TGF-β/Smad pathway mediates the dysfunction of NK cells and induces immune escape in B-acute lymphoblastic leukemia (ALL).10 Similar carcinogenic effects were also observed in renal cancer. For instance, Liu et al revealed that miR-93 targets RUNX3 and regulates the TGF-β/Smad4 signaling pathway to accelerate RCC.11 Interestingly, FAST-1 mediates embryogenesis by transducing TGF-β superfamily signals.12
In this study, we found FAST1 was significantly up-regulated in terms of its expression in RC tissues. Further analysis showed that a higher level of FAST1 was associated with lower survival of RC patients. Combined with the above findings, both FAST1 and the TGF-β/Smad signal can interfere with RC progression. Therefore, we hypothesized that FAST1 influences the progression of RC by regulating the TGF-β/Smad signaling pathway.
Patients and Methods
Clinical Samples
Twenty-eight patients who underwent surgical RCC in Zaozhuang Municipal Hospital from June 2016 to June 2017 were selected, and their RCC tissues and paired normal adjacent tissues were obtained. Tissue samples were immediately frozen in −80°C liquid nitrogen for subsequent experiments. All the sample patients did not receive radiotherapy, chemotherapy and other adjuvant treatment before the surgery. All samples were confirmed by the pathology department of our hospital. The patients and their family members signed informed consent. This study was authorized by the Ethics Committee of Zaozhuang Municipal Hospital. The FAST1 expression was tested by quantitative reverse transcription PCR (qRT-PCR), and the expression level > 2.5 was defined as high.
Cell Culture and Transfection
Human normal proximal renal tubular cell lines (OAT1) and RC cell lines (769-P and ACHN) were bought from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in the RPMI-1640 medium (Thermo Fisher Scientific, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, MA, USA) and 1% penicillin/streptomycin (Invitrogen, CA, USA) and incubated at 37 °C with 5% CO2. When the cell abundance reached 70%-80%, FAST1 overexpression plasmids (FAST1), knockdown plasmids (Si-FAST1#1 and Si-FAST1#2), and corresponding control group (NC and Si-NC) were transfected into 769-P and ACHN with the Lipofectamine2000 transfection agent (ThermoFisherScience, Waltham, MA, USA). The cells were collected for subsequent experiments after incubation for 48 hours. The TGF-β inhibitor RepSox was obtained from Abcam (2 µmol/L, ab142139, USA).
qRT-PCR
Total RNA was isolated from 769-P and ACHN cells with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and its concentration was measured with a NanoDrop2000 photometer (Thermo Fisher, Waltham, MA, USA). Then, RNA was reverse-transcribed into cDNA using the PrimeScript™ RT Reagent kit (Invitrogen, Shanghai, China) following the manufacturer’s instructions, and qRT-PCR was operated on the Applied Biosystems StepOnePlus System (Foster City, CA, USA). GAPDH was the internal reference of FAST1, and the 2(-ΔΔCt) method was adopted for statistics. Each experiment was done three times. The specific primer sequences include FAST1, forward, 5ʹ-GAGGTACCTGCGACATGACA-3ʹ, reverse, 5ʹ-GTTGTGGCGAATGGAGTCTT-3ʹ. GAPDH, forward, 5ʹ- TGGTTGAGCACAGGGTACTT −3ʹ reverse: 5ʹ- CCAAGGAGTAAGACCCCTGG −3ʹ.
Western Blot (WB)
The cells were lysed in the RIPA lysis buffer and electrically transferred to the polyvinylidene fluoride (PVDF) membranes, which were then blocked with 5% skimmed milk for 1 hour at room temperature (RT) and washed with TBST three times (10 min each time). Afterward, the membranes were incubated with the primary Anti-FAST1 antibody (1:1000, ab189960, Abcam, USA), Anti-E-Cadherin antibody (1:1000, ab231303, Abcam, USA), Anti-Claudin 1 antibody (1:1000, ab211737, Abcam, USA), Anti-N Cadherin antibody (1:1000, ab76057, Abcam, USA), Anti-Snail antibody (1:1000, ab82846, Abcam, USA), Anti-TGF-β (phospho S165) (1:1000, ab183037, Abcam, USA), Anti-TGF-β antibody (1:1000, ab36495, Abcam, USA), Anti-Smad2 (phospho S255) antibody (1:1000, ab188334, Abcam, USA), Anti-Smad2 antibody (1:1000, ab40855, Abcam, USA) and Anti-GAPDH antibody (1:1000, ab181603, Abcam, USA) at 4 °C overnight. After being rinsed with TBST, the membranes were incubated with the horseradish peroxidase (HRP)-labeled Goat Anti-Rabbit IgG (1:3000, ab6721, Abcam, USA) for 1 hour at RT. Then, the membranes were washed with TBST three times (10 min each time). Finally, ECL Western blot special reagents (Invitrogen) were used for color development and imaging, and Image J was applied to analyze each protein’s gray intensity.
Cell Colony Formation Experiment
The stably transfected 769-P and ACHN cells were inoculated in 6-well plates at 1×103/well and treated with overexpressed FAST1 and knocked down FAST1 to testify the colony formation. After the cells were incubated at 37 °C with 5% CO2 for two weeks, they were cleared with PBS and stained with crystal violet solution (0.1% crystal violet) for 15 minutes. The stained colonies were counted with an optical microscope (Olympus, Tokyo, Japan).
Flow Cytometry
Two groups of stably transfected cells were inoculated in 96-well plates at 1×104 cells/well. After 24 hours of culture, the cells were washed twice with PBS, fixed with 75% ethanol, and preserved overnight at 4 °C. The cells were then suspended in PBS washing buffer (1×106 cells/mL). Afterward, the propidium iodide staining solution was added for staining for 30 minutes, and the cells were stained with the Annexin V-fluorescein isothiocyanate/PI kit (cat. no. 556,570, BD Biosciences, New Jersey, USA). Flow cytometry (Beckman Coulter, Fullerton, CA, USA) was employed for cell detection, and FlowJo 10 (FlowJo LLC) was adopted for apoptosis analysis.
Cell Counting Kit-8 (CCK8) Assay
Two groups of cells (2×103 cells in each group) were taken and inoculated into 96-well plates. Cell proliferation was detected with the CCK8 kit (Beyotime Biotechnology, Shanghai, China), and 10 μL of CCK8 solution was added to each well after 24 hours of adherent culture. The culture plates were incubated for 4 hours, and the absorbance value was observed with a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) at 450 nm. The proliferation changes were monitored at 0, 24, 48, 72, and 96 hours after the transfection or the inhibitor treatment.
Transwell Assay
Transwell assay was employed to test cell migration and invasion. For the cell migration experiment, cells were seeded to the upper chamber of the transwell (2×104/well), and 200 μL culture medium containing 20% FBS was added to the lower chamber for culture at 37 °C. After 12 hours, the cells in the upper chamber were removed, fixed with 4% paraformaldehyde, stained with 0.1% crystal violet for 20 minutes, and washed with PBS. In the invasion assay, the transwell’s upper chamber was precoated with the matrigel (Biosciences, Lincoln, NE) before the cells were added, and the remaining steps were the same as that of the migration assay.
Animal Experiment
BALB/c nude mice (6–8 weeks old) were bought from the JunKeBiological Co., Ltd. (Nanjing, China). They were divided into five groups (five mice in each group) and raised in a specified pathogen-free environment. Meanwhile, 769-P cells stably transfected with FAST1 overexpression plasmids were trypsinized to make single-cell suspension (1×107 cells/mL). Then, 0.1 mL of the suspension was subcutaneously injected into both sides of the back of each nude mouse. The tumor size was measured with a caliper every four days from day 12. The volume was equal to the long diameter × the short diameter2/2. In the fourth week, mice were euthanized, and the tumors in nude mice were carefully dissected and weighed. These trials were conducted in strict accordance with the Guidelines for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH Publication No. 85–23, 2011). The Ethics Committee of Zaozhuang Municipal Hospital approved all experimental procedures.
Bioinformatic Analysis
Through STRING database (https://string-db.org/), the potential biological function of FAST1 was analyzed by gene ontology (GO) enrichment analysis.13 Next, the protein-protein interaction network was also constructed, which showed the functional interactions between proteins thus insighting into the molecular mechanism of cellular processing. The protein-protein interaction (PPI) network of FAST1 with other proteins were constructed by the online website of STRING. The interacting pairs with a combined score >0.7 (high confidence) were selected for the PPI network construction.
Co-Immunoprecipitation
The 769-P cells were cultured in 6-well plates with 5×105 cells each well. 24 hours after the incubation, the cells were transfected or co-transfected with plasmids, as shown in the figures for 24 h, the nuclear protein was extracted for immunoprecipitation. The ANTI-FLAG® M2 Affinity Gel was added into the lysis and incubated with continuing slow rotation at 4 °C overnight. After washing and denatured, the samples were used for Western blot analysis.
Immunohistochemistry
Immunohistochemistry was used to detect FAST1, p-SMAD2 and E-cadherin in the tumor tissues. 4-um thick histological sections of the paraffin-embedded tissues were prepared, dewaxed, deparaffinized in fresh xylene and dehydrated using a graded series of ethanol solutions. Next, the sections received antigen retrieval (in citrate buffer (0.01 M, PH 6.0)) and then incubation with 1% bovine serum albumin. After that, the sections were then incubated with primary antibodies (FOXH1, 1:200, Biorbyt; p-SMAD2, 1:200, abcam; E-cadherin, 1:500, abcam) at 4 °C overnight. At the next day, horseradish peroxidase-labelled anti-rabbit IgG was used to incubate with the sections after being washed with PBS. Finally, the stained images were observed under a light microscope (Olympus, Japan).
Statistical Analysis
GraphPad Prism 6 Software (GraphPad Software, Inc., City, State) was employed for analysis. The data were presented as mean ± standard deviation (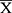 ±s). The multi-factor comparison was made by one-way ANOVA, while t-test was used for pairwise comparison. P<0.05 indicated statistical significance.
±s). The multi-factor comparison was made by one-way ANOVA, while t-test was used for pairwise comparison. P<0.05 indicated statistical significance.
Results
FAST1 Was Up-Regulated in RC
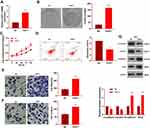
QRT-PCR and WB were carried out to testify the FAST1 level, and it was found that the FAST1 level in RC tissues was much higher than that in adjacent normal tissues (P < 0.05, Figure 1A and B). Additionally, the patients’ survival analysis manifested that patients with low FAST1 expression had a higher survival rate (P=0.043, Figure 1C). Through GEPIA (http://gepia.cancer-pku.cn/), we verified that the higher level of FAST1 in kidney renal clear cell carcinoma (KIRC) predicts the poorer survival rate (P < 0.05, Figure 1D). By analyzing the clinical characteristics of the sample patients, it was found that the higher expression of FAST1 was significantly associated with a more advanced stage of RC, indicated by the higher TNM stage and lymphatic metastasis rate (Table 1). These results indicated that FAST1 was overexpressed in RC and might be a new diagnostic marker of RC.
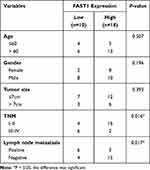 |
Table 1 Association Between Clinicopathological Features and FAST1 Expression in RCC Tissues (n =28) |
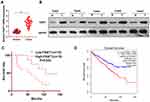 |
Figure 1 FAST1 was up-regulated in RC tissues. RC tissues and paired paracancerous tissues were harvested and stored in −80°C liquid nitrogen. (A and B) QRT-PCR and WB were applied to test the FAST1 expression in RC tissues and adjacent normal tissues, ***P < 0.001 (vs Normal group); (C) survival of RCC patients were analyzed via KM Plotter assay. (D) GEPIA (http://gepia.cancer-pku.cn/) was adopted to query the FAST1 expression and survival prognosis in KIRC. |
Overexpressing FAST1 Aggravated the Malignant Behaviors of RC Cells
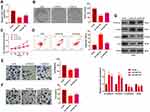
We constructed a FAST1 overexpression model in 769-P cell lines to probe the impact of FAST1 on RC development (P< 0.05, Figure 2A). Then, the colony formation assay and CCK8 experiment were conducted to test cell proliferation and viability. It was confirmed that overexpressing FAST1 significantly enhanced cell viability and proliferation (P < 0.05, Figure 2B and C). Flow cytometry demonstrated that the number of apoptotic cells was declined after overexpression of FAST1 (P < 0.05, Figure 2D). Transwell assay testified that cell migration and invasion were strengthened after overexpressing FAST1 (P < 0.05, Figure 2E and F). Moreover, WB was employed to monitor the epithelial-mesenchymal transition (EMT) markers (E-cadherin, Claudin1, N-cadherin, and Snail), and it was found that the expression of E-cadherin and Claudin1 was higher, while the expression of N-cadherin and Snail was lower in the FAST1 group than that of the control group (P < 0.05, Figure 2G). These results suggested that FAST1 overexpression expedited RC and might be an oncogene of RC.
Knockdown of FAST1 Repressed the Malignant Biological Behaviors of RC and Induced Apoptosis
We constructed a FAST1 knockdown model in ACHN cell lines to verify the role of FAST1 in RC (P < 0.05, Figure 3A). Then, cell proliferation, viability, apoptosis, migration, and invasion were determined by the colony formation experiment, flow cytometry, CCK8 and Transwell assay, respectively. The results confirmed that knocking down FAST1 weakened cell viability and proliferation, promoted apoptosis, and abated cell migration and invasion (P < 0.05, Figure 3B–F). Also, WB showed that compared with the control group, E-cadherin and Claudin1 expression was increased, while the N-cadherin and Snail expression was abated in the Si-FAST1 group (P < 0.05, Figure 3G). These findings illustrated that knocking down FAST1 inhibited the malignant biological behaviors of RC cells and prevented EMT.
FAST1 Promoted the Growth of RC Cells in vivo
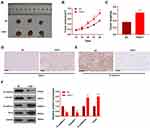
Furthermore, 769-P cells transfected with FAST1 overexpression plasmids were injected into nude mice, and the tumor growth was observed in vivo. The tumor size was determined from day 12 and recorded every four days, and a growth curve was plotted. As a result, the mice’s tumor volume and weight were significantly increased after overexpressing FAST1 compared with the control group (P < 0.05, Figure 4A–C). We then performed IHC to detect FAST1 in the tumor tissues. It was found that compared with the NC group, FAST1 was overexpressed in the tumor tissues, mainly located at the cytoplasm and nucleus (Figure 4D). Then, the EMT marker expression in the tumor was tested by IHC and WB. The results confirmed that the levels of E-cadherin and Claudin1 in the FAST1 group were higher than those in the control group, while the profiles of N-cadherin and Snail were completely opposite (P < 0.05, Figure 4E and F). Thus, FAST1 served as an oncogene in vivo, promoting tumor growth and EMT.
FAST1 Induced the TGF-β/Smad2 Signaling Activation
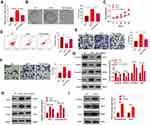
To clarify the specific mechanism of the carcinogenic function of FAST1 in RC, we investigated its downstream TGF-β/Smad2 pathway. The levels of TGF-β, Smad2 in RC cells were determined by WB, and it was discovered that their expression was higher in the FAST1 overexpression group than that in the control group. In contrast, knocking down FAST1 inhibited TGF-β and p-Smad2 levels (P < 0.05, Figure 5A and B). Interestingly, TGF-β and p-Smad2 levels in the xenograft tumors were also increased with FAST1 up-regulation (Figure 5C and D). Additionally, we predicted the protein-protein interaction network of FAST1 and the downstream protein pathways through STRING (https://string-db.org/cgi/input.pl). The analysis result showed that FOXH1 functions as a transcriptional activator, recognizes and binds to the DNA sequence 5ʹ-TGT[GT][GT]ATT-3ʹ, is required for induction of the goosecoid (GSC) promoter by TGF-beta or activin signaling. Moreover, FOXH1 forms a transcriptionally active complex containing FOXH1/SMAD2/SMAD4 on a site on the GSC promoter called TARE (TGF-beta/activin response element) (Figure 5E). Moreover, FAST1 has potential function in regulating the TGF-beta signaling pathway (Figure 5F). To confirm the binding relationship between SMAD2 and FAST1, we conducted co-immunoprecipitation, the results showed that FAST1 formed a complex with the Smad2 (Figure 5G), suggesting that there was crosstalk between the FAST1 and Smad2. Overall, these results testified that FAST1 activated TGF-β/Smad2 pathway by binding with SMAD2.
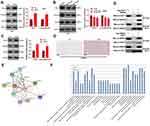 |
Figure 5 FAST1 activated TGF-β/Smad. The 789-P cell lines stably transfected with overexpressing FAST1, ACHN cell lines stably transfected with knocking down FAST1, and mouse tumors obtained by tumor formation experiment in nude mice were selected. (A–C) WB was employed to examine the expression of TGF-β and p-Smad2/Smad2 in 789-P or ACHN cells or in vivo tumor tissues. (D) IHC was performed to detect p-Smad2 expression in the tumor tissues. (E) STRING (https://string-db.org/cgi/input.pl) showed the protein protein interaction (PPI) network of FAST1. (F) The downstream protein pathways of FAST1 were predicted through STRING (https://string-db.org/cgi/input.pl), TGF-β signaling pathway was potential pathways underlying FAST1. (G) Co-immunoprecipitation was used to confirm the interaction between FAST1 and Smad2 in 789-P cells. **P < 0.01, ***P < 0.001 (vs NC group); *P < 0.05, **P < 0.01, ***P < 0.001 (vs Si-NC group). N=5. |
Inhibiting TGF-β Attenuated Malignant Biological Behaviors of RC Induced by Overexpressing FAST1
We further investigated whether TGF-β affected the effect of FAST1 on RC. The TGF-β inhibitor (RepSox) was added on the basis of FAST1 overexpression, and the FAST1 level was monitored by qRT-PCR. Interestingly, the FAST1 expression was higher after overexpressing FAST1 (vs the control group), and its level was lower in the FAST1+RepSox group than that in the FAST1 group (P < 0.05, Figure 6A). Next, the cell viability, apoptosis, migration and invasion were measured by colony formation, flow cytometry, CCK8 and Transwell assay, respectively. Interestingly, the FAST1+RepSox group had eliminated cell proliferation, migration and invasion, and had enhanced apoptosis compared with FAST1 group (P < 0.05, Figure 6B–F). Additionally, WB results manifested that FAST1+RepSox inhibited the EMT induced by overexpressing FAST1, indicated by up-regulated E-cadherin and Claudin1 and down-regulated N-cadherin and Snail (Figure 6G). At the same time, FAST1+RepSox attenuated the activation of FAST1 on the TGF-β/Smad2 signaling pathway. Namely, TGF-β and p-Smad2/Smad2 were down-regulated (P < 0.05, Figure 6H). Moreover, we treated 769-P cells with TGF-β. The result of Western blot showed that TGF-β markedly enhanced p-Smad2 level, and also promoted FAST1 expression (Figure 6I). The above results proved that inhibiting TGF-β reduced the oncogenic effect of FAST1 in RC.
Discussion
RCC is the most common type of RC with a high degree of malignancy. According to statistics, there were about 627,000 new RCC cases in the United States in 2016, and the current incidence is still on the rise.14 Although the popularity of new technologies has improved the diagnostic rate of RC to some extent, the specific markers related to RC are still lacking. In this study, we found the potential of FAST1 as a biomarker for RC diagnosis and prognosis. Namely, FAST1 is highly expressed in RC tissues and is related to patients’ adverse prognosis, and knocking down FAST1 inhibits the RC evolvement.
Earlier studies have revealed that Forkhead box transcription factors can regulate cell proliferation, apoptosis, migration, metabolism and DNA damage response.15 For example, Forkhead Box M1 (FOXM1), which is up-regulated in malignant meningioma tissues, facilitates meningioma cell viability, proliferation, tube formation and inhibits apoptosis by repressing AHR, CYP1A1 and VEGFA.16 Interestingly, several Forkhead box transcription factors such as FOXM117 and FOXA118 are found to induce oncogenic effects in RC, while the other Forkhead box transcription factor members, including FOXO3,19 FOXL120 and FOXO1,21 show potent anti-tumor effects by mitigating the proliferation and invasion of RC cells. As a novel member of the Forkhead box transcription factor family, FOXH1 has been revealed to modulate the malignant behaviors of tumor cells.5,6 Recently, Liu Y et al reported that FOXH1 significantly promotes cell proliferation, invasion and tumorigenesis of breast cancer cells. Benzyl isothiocyanate (BITC), an anti-tumor reagent in breast cancer, can exert a magnified tumor-suppressive effect when FOXH1 is down-regulated.22 In the present study, FAST1 was found significantly up-regulated in RC and associated with poorer survival or RC patients. Therefore, it is reasonable to speculate that FAST1 may serve as an oncogene in RC. Our experimental results confirmed that forced FAST1 overexpression promotes RC cell proliferation, viability, invasion, migration, and inhibits apoptosis rate. Therefore, FAST1 is a potential biological target for RC diagnosis and treatment.
EMT refers to the process that epithelial cells lose the apical-basal polarity and cell–cell adhesion and transit to invasive mesenchymal cells.23 In terms of its functions, EMT plays a vital role in modulating embryonic development, wound healing, cancer cell metastasis, and drug resistance.24–27 It is worth noting that in non-small cell lung cancer (NSCLC), UBE2T enhances the degradation of FOXO1 in a ubiquitination-dependent manner, thus repressing EMT of NSCLC cells.28 Similarly, this study found that FAST1 up-regulation led to up-regulated N-cadherin and Snail but inhibited E-cadherin and Claudin1, indicating FAST1 promoted the EMT of RC cells. Moreover, down-regulation of FAST1 led to the reduced potential of EMT in RC. As a result, we concluded that FAST1 aggravated the migration and invasion by promoting EMT of RC cells.
A previous study has found that FAST1 can interact with the Smad2 pathway, thereby mediating biological processes.29 TGF-β is a highly effective cytokine that contributes to maintaining cell homeostasis, embryonic development, and cell differentiation. It is considered an effective cell growth inhibitor. Above all, tumor cells can antagonize the inhibition of TGF-β in advanced stages, resulting in the conversion of TGF-β into cancer promoters.8 It has been found in the literature that TGF-β functions through its upstream and downstream molecular interactions, and Smad is one of the classic TGF-β dependent proteins. When TGF-β ligand binds to TGF-β1 and TGF-β2 receptors, TGF-β1 and TGF-β2 form a heterologous complex, which directly activates Smad2 and Smad3. While Smad2, 3 and 4 form trimers and transfer to the nucleus, regulating the target gene transcription through synergistic action.30,31 The TGF-β/Smad signal is involved in the development of multiple malignancies. For instance, Zhang et al showed that overexpressed SALT4 promotes the metastasis of gastric cancer by activating the TGF-β/Smad signaling pathway.32 Also, Gao et al claimed that Topoisomerase II (TOP2A) aggravates the progression of advanced ovarian cancer by activating TGF-β/Smad,33 suggesting that inhibiting the TGF-β/Smad signaling pathway may be a novel method for cancer treatment. Some scholars have found that the activation of down-regulated miR-542-3p on the TGF-β/Smad signaling pathway mediates hepatocellular carcinoma progression.34 In addition, Dong et al reported that the MMP-9 inhibitor effectively blocked the activation of TGF-β/Smad, thus inhibiting the invasion of breast cancer.35 Fortunately, TGF-β/Smads activation is demonstrated to play an oncogenic role in RCC.36 In our study, we found that FAST1 positively regulates the TGF-β/SMAD pathway. Importantly, the TGF-β inhibition partly reversed the FAST1-mediated promoting effect in RC. Specifically, FAST1 potentials RC development dependently through the TGF-β pathway.
Overall, FAST1 is up-regulated in RC and also promotes the progression of RC. This pro-oncogenic effect of FAST1 in RC is closely related to the activation of its downstream TGF-β/Smad signaling pathway. Nevertheless, the sample size involved in this paper is small and cannot represent a large range of the population. It is necessary to further expand the sample source and diversity to testify its applicability in clinical practice.
Data Sharing Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
Our study was approved by the Ethics Review Board of Zaozhuang Municipal Hospital.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not- for- profit sectors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang AB, Bao Y, Wu ZJ, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10(3):154. doi:10.1038/s41419-019-1331-9
2. Liu XB, Hao YX, Yu W, et al. Long non-coding RNA emergence during renal cell carcinoma tumorigenesis. Cell Physiol Biochem. 2018;47(2):735–746. doi:10.1159/000490026
3. Wang L, Su Y, Huang C, et al. FOXH1 is regulated by NANOG and LIN28 for early-stage reprogramming. Sci Rep. 2019;9(1):16443. doi:10.1038/s41598-019-52861-8
4. Roessler E, Ouspenskaia MV, Karkera JD, et al. Reduced NODAL signaling strength via mutation of several pathway members, including FOXH1, is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83(1):18–29. doi:10.1016/j.ajhg.2008.05.012
5. Cheng Q, Shi YJ, Li Z, Kang H, Xiang Z, Kong LF. FAST1 promotes the migration and invasion of colorectal cancer cells. Biochem Biophys Res Commun. 2019;509(2):407–413. doi:10.1016/j.bbrc.2018.12.122
6. Huang CX, Chen N, Wu XJ, et al. Zebrafish let-7b acts downstream of hypoxia-inducible factor-1α to assist in hypoxia-mediated cell proliferation and cell cycle regulation. Life Sci. 2017;171:21–29. doi:10.1016/j.lfs.2017.01.005
7. Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–978. doi:10.7150/ijbs.4564
8. Oshimori N, Oristian D, Fuchs E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160(5):963–976. doi:10.1016/j.cell.2015.01.043
9. Massagué J, Seoane J, Wotton D, et al. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi:10.1101/gad.1350705
10. Rouce RH, Shaim H, Sekine T, et al. The TGF-β/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia. 2016;30(4):800–811. doi:10.1038/leu.2015.327
11. Liu LJ, Yu JJ, Xu XL. MicroRNA-93 inhibits apoptosis and promotes proliferation, invasion and migration of renal cell carcinoma ACHN cells via the TGF-β/Smad signaling pathway by targeting RUNX3. Am J Transl Res. 2017;9(7):3499–3513.
12. Weisberg E, Winnier GE, Chen X, Farnsworth CL, Hogan BL, Whitman M. A mouse homologue of FAST-1 transduces TGF beta superfamily signals and is expressed during early embryogenesis. Mech Dev. 1998;79(1–2):17–27. doi:10.1016/S0925-4773(98)00160-9
13. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Databaseissue):D808–D815. doi:10.1093/nar/gks1094
14. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
15. Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi:10.1016/j.lfs.2015.12.001
16. Wu Q, Zheng J, Dou C, Qi S. Forkhead box M1 promotes the growth and tube formation of human malignant meningioma cells via the aryl hydrocarbon receptor signaling pathway. Folia Neuropathol. 2020;58(3):223–236. doi:10.5114/fn.2020.100065
17. Liu F, Li N, Liu Y, Zhang J, Zhang J, Wang Z. Homeodomain interacting protein kinase-2 phosphorylates FOXM1 and promotes FOXM1-mediated tumor growth in renal cell carcinoma. J Cell Biochem. 2019;120(6):10391–10401. doi:10.1002/jcb.28323
18. Tong Z, Meng X, Wang J, Wang L. MicroRNA-212 inhibits the proliferation and invasion of human renal cell carcinoma by targeting FOXA1. Mol Med Rep. 2018;17(1):1361–1367. doi:10.3892/mmr.2017.7956
19. Nie W, Ni D, Ma X, et al. miR-122 promotes proliferation and invasion of clear cell renal cell carcinoma by suppressing Forkhead box O3 [published correction appears. Int J Oncol. 2019;54(2):559–571. doi:10.3892/ijo.2018.4636
20. Yang FQ, Yang FP, Li W, et al. Foxl1 inhibits tumor invasion and predicts outcome in human renal cancer. Int J Clin Exp Pathol. 2013;7(1):110–122.
21. Zhou L, Yin B, Liu Y, Hong Y, Zhang C, Fan J. Mechanism and function of decreased FOXO1 in renal cell carcinoma. J Surg Oncol. 2012;105(8):841–847. doi:10.1002/jso.23013
22. Liu Y, Zhang L, Meng Y, Huang L. Benzyl isothiocyanate inhibits breast cancer cell tumorigenesis via repression of the FoxH1-mediated Wnt/β-catenin pathway. Int J Clin Exp Med. 2015;8(10):17601–17611.
23. Zhao Y, Ye X, Chen R, et al. Sirtuin 7 promotes non-small cell lung cancer progression by facilitating G1/S phase and epithelial-mesenchymal transition and activating AKT and ERK1/2 signaling. Oncol Rep. 2020;44(3):959–972. doi:10.3892/or.2020.7672
24. Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21(7):965. doi:10.3390/molecules21070965
25. Dong J, Zhai B, Sun W, Hu F, Cheng H, Xu J. Activation of phosphatidylinositol 3-kinase/AKT/snail signaling pathway contributes to epithelial-mesenchymal transition-induced multi-drug resistance to sorafenib in hepatocellular carcinoma cells. PLoS One. 2017;12(9):e0185088. doi:10.1371/journal.pone.0185088
26. Navas T, Pfister TD, Colantonio S, et al. Novel antibody reagents for characterization of drug- and tumor microenvironment-induced changes in epithelial-mesenchymal transition and cancer stem cells. PLoS One. 2018;13(6):e0199361. doi:10.1371/journal.pone.0199361
27. Wang H, Zhang B, Wang X, et al. TRPV4 overexpression promotes metastasis through epithelial-mesenchymal transition in gastric cancer and correlates with poor prognosis. Onco Targets Ther. 2020;13:8383–8394. doi:10.2147/OTT.S256918
28. Yin H, Wang X, Zhang X, et al. UBE2T promotes radiation resistance in non-small cell lung cancer via inducing epithelial-mesenchymal transition and the ubiquitination-mediated FOXO1 degradation. Cancer Lett. 2020;494:121–131. doi:10.1016/j.canlet.2020.06.005
29. Noda D, Itoh S, Watanabe Y, et al. ELAC2, a putative prostate cancer susceptibility gene product, potentiates TGF-beta/Smad-induced growth arrest of prostate cells. Oncogene. 2006;25(41):5591–5600. doi:10.1038/sj.onc.1209571
30. Yoshimatsu Y, Wakabayashi I, Kimuro S, et al. TNF-α enhances TGF-β-induced endothelial-to-mesenchymal transition via TGF-β signal augmentation. Cancer Sci. 2020;111(7):2385–2399. doi:10.1111/cas.14455
31. Nakao A, Imamura T, Souchelnytskyi S, et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16(17):5353–5362. doi:10.1093/emboj/16.17.5353
32. Zhang X, Zhang P, Shao M, et al. SALL4 activates TGF-β/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag Res. 2018;10:4459–4470. doi:10.2147/CMAR.S177373
33. Gao Y, Zhao HY, Ren M, et al. TOP2A promotes tumorigenesis of high-grade serous ovarian cancer by regulating the TGF-β/Smad pathway. J Cancer. 2020;11(14):4181–4192. doi:10.7150/jca.42736
34. Zhang T, Liu W, Meng W, et al. down-regulation of miR-542-3p promotes cancer metastasis through activating TGF-β/Smad signaling in hepatocellular carcinoma. Onco Targets Ther. 2018;11:1929–1939. doi:10.2147/OTT.S154416
35. Dong HD, Diao HX, Zhao Y, et al. Overexpression of matrix metalloproteinase-9 in breast cancer cell lines remarkably increases the cell malignancy largely via activation of transforming growth factor-beta/SMAD signalling. Cell Prolif. 2019;52(5):e12633. doi:10.1111/cpr.12633
36. Tretbar S, Krausbeck P, Müller A, et al. TGF-β inducible epithelial-to-mesenchymal transition in renal cell carcinoma. Oncotarget. 2019;10(15):1507–1524. doi:10.18632/oncotarget.26682
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.