Back to Journals » Clinical Epidemiology » Volume 10
Familial aggregation and heritability of type 1 diabetes mellitus and coaggregation of chronic diseases in affected families
Authors Kuo CF , Chou IJ , Grainge MJ , Luo SF, See LC , Yu KH, Zhang W , Doherty M, Valdes AM
Received 25 April 2018
Accepted for publication 26 June 2018
Published 10 October 2018 Volume 2018:10 Pages 1447—1455
DOI https://doi.org/10.2147/CLEP.S172207
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Henrik Sørensen
Chang-Fu Kuo,1,2,* I-Jun Chou,3,4,* Matthew J Grainge,5 Shue-Fen Luo,2 Lai-Chu See,2,6,7 Kuang-Hui Yu,2 Weiya Zhang,1 Michael Doherty,1 Ana M Valdes1
1Division or Rheumatology, Orthopaedics and Dermatology, School of Medicine, University of Nottingham, Nottingham, UK; 2Division of Rheumatology, Allergy and Immunology, Chang Gung Memorial Hospital, Taoyuan, Taiwan; 3Division of Clinical Neuroscience, School of Medicine, University of Nottingham, Nottingham, UK; 4Division of Paediatric Neurology, Chang Gung Memorial Hospital, Taoyuan, Taiwan; 5Division of Epidemiology and Public Health, School of Medicine, University of Nottingham, Nottingham, UK; 6Biostatistics Core Laboratory, Molecular Medicine Research Centre, Chang Gung University, Taoyuan, Taiwan; 7Department of Public Health, College of Medicine, Chang Gung University, Taoyuan, Taiwan
*These authors contributed equally to this work
Purpose: To estimate the extent of familial aggregation of type 1 diabetes (T1D) and coaggregation of related chronic diseases and assess the relative contribution of environmental and genetic factors on the risks.
Patients and methods: This population-based study used the Taiwan National Health Insurance database to reconstruct family structure and identify people with T1D and other chronic diseases between 1999 and 2015. Relative risks (RRs) for T1D and other chronic diseases and heritability of T1D were estimated. Heritability was estimated using the polygenic liability model.
Results: Validation of family structure found the positive predictive value to be 98.7% for maternal links and 98.6% for paternal links. Having an affected twin, first-degree relative, or spouse was associated with an adjusted RR (95% CI) of 553.66 (427.59–716.89), 32.49 (28.66–36.84), and 2.17 (0.31–15.40) for T1D, respectively. Based on the polygenic liability model, heritability, shared and non-shared contributions to T1D, and variances were 66.50%, 10.86%, and 22.64%, respectively. A family history of T1D was associated with an RR (95% CI) of 1.51 (1.20–1.89) for rheumatoid arthritis, 1.66 (1.21–2.26) for Sjögren’s syndrome, 1.48 (1.09–2.01) for systemic lupus erythematosus, 1.24 (1.14–1.35) for simple goiter, 1.16 (1.04–1.31) for non-toxic nodular goiter, 1.61 (1.49–1.74) for thyrotoxicosis, 1.78 (1.57–2.01) for acquired hypothyroidism, 1.66 (1.40–1.98) for thyroiditis, and 1.15 (0.97–1.37) for epilepsy.
Conclusion: These data highlight the importance of the genetic contribution to T1D and confirm the coaggregation of autoimmune and metabolic diseases with T1D.
Keywords: type 1 diabetes mellitus, familial aggregation, co-aggregation, autoimmune diseases, endocrine diseases
Introduction
Type 1 diabetes mellitus is an autoimmune disorder caused by the destruction of pancreatic β cells in people with a genetic predisposition, and it is characterized by absolute insulin deficiency.1,2 Although type 1 diabetes has a relatively low prevalence, epidemiologic studies have reported that the incidence of type 1 diabetes is increasing by 3% per year worldwide.3 It is one of the most common autoimmune diseases in childhood with an estimated incidence rate of 6.4/100,000 for 0- to 9-year-old Asians and Pacific Islanders.4 A recent population-based estimate of the incidence of type 1 diabetes in Taiwan was 5.6/1,000,000 for 0- to 14-year-olds,5 with a two- to three-fold higher mortality rate than age-matched individuals.6
Positive family history is one of the strongest risk factors for type 1 diabetes, and the genetic contribution to the susceptibility to type 1 diabetes is substantial.7 For example, the human leukocyte antigen (HLA) region has been found to be one of the major genetic determinants of type 1 diabetes.1,8 Strong ethnic differences in the HLA haplotypes present in this region,9 such as those between European and East Asian populations,10 are likely to explain some of the large variations in the prevalence and incidence of type 1 diabetes. To date, heritability estimates for type 1 diabetes have been derived only for populations of European origin,11,12 and the results have suggested an additive genetic contribution of 72%–88%. The heritability in East Asian populations has not been investigated to date. In addition, it is well known that type 1 diabetes coaggregates with other autoimmune diseases such as autoimmune thyroid disease and celiac disease.13 Recently, Chou et al reported that individuals with type 1 diabetes were more likely to develop epilepsy than controls,14 a finding that was later confirmed in a European cohort.15 Therefore, there may be a common root for type 1 diabetes with these chronic conditions.
This nationwide study used data on the entire population of Taiwan in 2015 to construct nationwide genealogy linking to health information derived from a large comprehensive database. We aimed to estimate the relative risks (RRs) of type 1 diabetes in individuals with affected relatives of specific kinship, and assess the relative contribution of genetic, and shared and non-shared environmental factors to the susceptibility to type 1 diabetes. In addition, we estimated the RRs of other chronic diseases in people with a family history of type 1 diabetes.
Materials and methods
Study population
Ethical approval was obtained from the Institutional Review Board of Chang Gung Memorial Hospital and the National Health Research Institute (IRB No 104-8042B), the data holder of the National Health Insurance (NHI) database. The NHI database was provided by the Administration of National Health Insurance, Ministry of Health and Welfare. The database is fully anonymized, and no trackable data were available. To protect confidentiality, the conduct of the research and use of the data within the database are restricted to the specialized data center operated by the administration. We constructed a cohort comprising all beneficiaries of the Taiwan NHI program who were registered in 2015 and had valid insurance status using data from the Registry for NHI beneficiaries.
The Taiwan NHI program was initiated in 1995 as a single-payer insurance system co-funded by the government, employers, and beneficiaries. As a single-payer system and as enrollment is required by law,16 almost all residents in Taiwan are enrolled, and the majority of medical facilities in Taiwan are contracted with the NHI to provide a comprehensive package of health care. The NHI database contains all registration and original claims data, including gender, date of birth, place of residence, details of insurance, family relationships, dates of inpatient and outpatient visits, medical diagnoses, medical expenditure, prescription details, examinations, operations, and procedures. All information in the database and other government-held data such as the death registry are linked using a unique personal identifier assigned to each resident in Taiwan. The personal identifier is encrypted; however, the identification remains unique for each beneficiary in the database to facilitate internal and external linkage of records.
Methods of nationwide genealogy reconstruction
We have reported methods for genealogy reconstruction using family relationships recorded in the NHI database in our previous publications.17–19 The registry of beneficiaries records the insurance status of all people covered by the NHI program, and also contains information regarding relationships between the insured person (who paid the insurance fee) and his/her dependents. Both blood relatives and spouses are eligible as dependents of the insured. A birth certificate is required for a child to be registered as a dependent of their parents. The family identifiers and unique personal identifiers allowed us to establish family relationships (parents, offspring, full siblings, twins, and spouse). We used an algorithm to identify parent–offspring relationships indirectly and to maximize possible family links.17–19 Full siblings were defined as those with the same parents. Twins were defined as full siblings with the same date of birth (± 1 day). However, twin zygosity could not be derived from the database. Changes in residence, employment, insurance status (insurant or dependent), and relationships between the insurant and dependents are recorded in the registry. Overall, there were ~364 million records in the registry in 2015. We used this dynamic nature to maximize the identification of possible family links by incorporating the entire registry of records from 1999 to 2015. We used family relationships to reconstruct genealogy. Overall, we classified 4,373,116 families of 2–5 generations with a mean family size of 5.8 persons. Each may have appeared multiple times in different categories of family relationships depending on the family structure.
Validation of family relationships
To validate the accuracy of the family identification method, we linked the NHI data to the civil registration system governed by the Ministry of Internal Affairs, which records the original data of the birth certificate. We identified 2,171,765 children born between 2001 and 2014 and found that the maternal and paternal links identified by our methods were highly consistent with the original birth certificate records. The positive predictive value was 98.7% for maternal links and 98.6% for paternal links.
Ascertainment of type 1 diabetes and other autoimmune diseases
In Taiwan, patients with suspected autoimmune diseases and severe endocrine diseases are referred to specialists for diagnosis and treatment. Patients with diagnoses of type 1 diabetes and other autoimmune diseases are entitled to a waiver for medical co-payments. Clinical information is sent to the insurance administration for review by commissioned expert panels to confirm the diagnosis before approving such waivers. The approval of co-payment waivers for type 1 diabetes requires further confirmation of the diagnosis such as the presence of autoantibodies. The Registry for Catastrophic Illness Patients contains clinical and registration information of these patients. We used this registry to identify patients with type 1 diabetes mellitus (ICD-9 codes 250.01, 250.03, 250.11, 250.13, 250.21, 250.23, 250.31, 250.33, 250.41, 250.43, 250.51, 250.53, 250.61, 250.63, 250.71, 250.73, 250.81, 250.83, 250.91, and 250.93). Therefore, all patients who had type 1 diabetes between 1999 and 2015 and were alive in 2015 were identified. The diagnosis of diabetes in the NHI database has previously been validated, with a positive predictive value of type 1 diabetes of 98.3%.6,20 The case definition of type 1 diabetes was also validated by a retrospective chart review.
The details of case definitions for other chronic conditions are shown in Table S1, and included rheumatoid arthritis, Sjögren’s syndrome, systemic lupus erythematosus, systemic sclerosis, polymyositis/dermatomyositis, Behçet’s disease, vasculitis, inflammatory bowel disease, multiple sclerosis, myasthenia gravis, simple goiter, non-toxic nodular goiter, thyrotoxicosis, acquired hypothyroidism, thyroiditis, and epilepsy.
Covariates
We considered that age, sex, occupation, income, level of urbanization of residence, and family size may have confounded or modified the familial associations. Therefore, these factors were used as covariates in the estimation of familial RRs.
Statistical analysis
The prevalence of type 1 diabetes for the general population was calculated as the number of people who were alive and diagnosed with type 1 diabetes between 1999 and 2015 divided by the number of mid-year population in Taiwan in 2015. This prevalence estimate was the basis for the calculations of heritability and familial transmission. The RRs of type 1 diabetes were the adjusted prevalence ratios between first-degree relatives of an individual with type 1 diabetes and the general population.21 Of the several established methods available for the estimation of prevalence ratios, we used Breslow–Cox models to estimate prevalence rate ratios in a cross-sectional study by setting an equal follow-up time for all subjects.22 This method has been shown to produce consistent estimates of prevalence ratios close to true parameters.23,24 The Breslow–Cox model assumes independence between subjects; however, family members form natural clusters. Bias caused by within-family clustering can be overcome by using the marginal model or shared frailty model.25 Previous studies have documented the comparability of point estimates and 95% CIs between the frailty model25 and marginal model with a robust sandwich method.26 In addition, a previous study suggested that the marginal model produces a more precise parameter.27 The RRs were adjusted for age, sex, socioeconomic factors, and family size.28 We fitted the models separately according to the kinship and sex of the affected relatives. Twins were excluded from the sibling analyses. The RR was estimated for the number of affected first-degree kinships. In this model, we compared the risk of type 1 diabetes in individuals with one or two affected first-degree relatives with the general population.
Based on the polygenic liability model,17–19,29 heritability was defined as the proportion of phenotypic variance attributable to genetic factors, and familial transmission was defined as the sum of genetic and shared environmental contributions. Familial transmission and heritability can be calculated using the polygenic liability model to calculate both measures.30–32 As in the original formula developed by Reich and Yang,29 we used the population prevalence and sibling RR to calculate familial transmission. We further used spouses as controls, assuming that they shared the family environment but had no genetic similarity with blood family members. We could thus calculate heritability, as reported in our previous publications.17–19
The extent of familial coaggregation with other chronic diseases in the families affected by type 1 diabetes was estimated using a marginal Cox proportional hazards regression model with an equal follow-up time for all subjects adjusting for age, sex, place of residence, income level, occupation, and family size. RRs were estimated as the adjusted prevalence ratio of the specified autoimmune disease between individuals with a first-degree relative with type 1 diabetes mellitus and those without a family history of type 1 diabetes mellitus. A two-sided P-value of less than 0.05 was considered to indicate statistical significance. All analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC, USA).
Results
The prevalence of type 1 diabetes in individuals with affected first-degree family members versus the general population
We identified 10,723 patients with a diagnosis of type 1 diabetes, giving a crude prevalence of 0.04% (Table 1). Women had a slightly higher prevalence than men. In the general population of Taiwan in 2015, 28,163 (0.12%) individuals had at least one first-degree relative with type 1 diabetes, including 4,872 with affected parents, 12,493 with affected offspring, 10,755 with an affected sibling, and 123 with an affected twin. The age-specific prevalence of type 1 diabetes was higher in the individuals with affected first-degree relatives with type 1 diabetes than in the general population (Figure 1).
  | Table 1 Baseline characteristics of the individuals with affected first-degree relatives with type 1 diabetes and the general population |
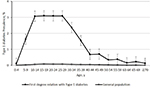  | Figure 1 Age-specific prevalence of type 1 diabetes in individuals with an affected first-degree relative and the general population in Taiwan in 2015. |
Relative risks for type 1 diabetes in individuals with affected first-degree relatives
The prevalence of type 1 diabetes in individuals with affected first-degree relatives of specific types is shown in Table 2. The RRs (95% CIs) for type 1 diabetes were associated with the degree of genetic distance between family relatives. Overall, having an affected co-twin (50% or 100% genetic similarity depending on zygosity), first-degree relative (on average 50% genetic similarity), or spouse (no genetic similarity) was associated with adjusted RRs of 553.66 (427.59–716.89), 32.49 (28.66–36.84), and 2.17 (0.31–15.40), respectively. In addition, the RRs increased with the number of types of affected first-degree relatives. The individuals with one type of affected first-degree relative had an RR (95% CI) of 31.19 (27.45–35.44), and those with two or more types of affected first-degree relatives had an RR of 173.84 (95.63–316.01) for type 1 diabetes.
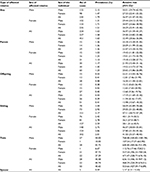  | Table 2 Relative risks for type 1 diabetes in first-degree relatives Note: aAdjusted for age, gender, place of residence, quintile of income level, occupation, and family size. |
Familial resemblance and heritability of type 1 diabetes
Using a threshold liability model, we estimated that the accountability for phenotypic variance in type 1 diabetes was 66.50% for genetic factors (heritability), 10.86% for shared environmental factors, and 22.64% for non-shared environmental factors.
Coaggregation of other autoimmune diseases, thyroid diseases, and epilepsy
We examined the RRs of ten other autoimmune diseases (Table 3), three of which (rheumatoid arthritis, Sjögren’s syndrome, and systemic lupus erythematosus) showed a significant familial coaggregation associated with type 1 diabetes. A family history of type 1 diabetes was associated with thyroid diseases, specifically, a RR (95% CI) of 1.24 (1.14–1.35) for simple goiter, 1.16 (1.04–1.31) for non-toxic nodular goiter, 1.61 (1.49–1.74) for thyrotoxicosis, 1.78 (1.57–2.01) for acquired hypothyroidism, and 1.66 (1.40–1.98) for thyroiditis. We further tested the coaggregation of type 1 diabetes with epilepsy, as an association was reported in a recent study. However, we found that the RR of epilepsy was not significantly increased (RR, 1.15; 95% CI, 0.97–1.37) in relatives of people with type 1 diabetes compared to the general population (Table 3).
  | Table 3 Relative risks of other diseases in first-degree relatives of patients with type 1 diabetes |
Discussion
This is the first study to report the heritability of type 1 diabetes in an East Asian population. Type 1 diabetes was more common in people with a family history, and the risk was associated with the genetic distance of affected relatives. The familial RR was 30 times higher than that of individuals with an affected first-degree relative compared to the general population. We also confirmed the strong genetic contribution to type 1 diabetes with a heritability of 66.5%, and our results showed that one-third of phenotypic variance was attributable to environmental factors. Furthermore, autoimmune diseases such as rheumatoid arthritis and thyroid diseases were more common in people with a family history of type 1 diabetes, suggesting that type 1 diabetes has a strong genetic association or shares common pathogenetic roots with autoimmune and thyroid diseases.
Several previous studies have investigated the heritability of type 1 diabetes in northern Europe,11,12 with heritability estimates ranging from 70% to 88%, which is slightly higher than our estimate of 66.5%. A key difference is that we found that shared environmental factors also contributed to phenotypic variance, whereas no effect was estimated by Hyttinen et al.11 We also found significant coaggregation with three autoimmune diseases, rheumatoid arthritis, Sjögren’s syndrome, and systemic lupus erythematosus, and also a trend toward an increased risk of systemic sclerosis, polymyositis/dermatomyositis, Behçet’s disease, vasculitis, and myasthenia gravis in the relatives of the patients with type 1 diabetes. The very low prevalence of these disorders in Taiwan means that the study may not be powered sufficiently to detect significant coaggregation with these disorders. However, a previous study found familial coaggregation of type 1 diabetes with autoimmune thyroiditis and celiac disease.33 Our data also showed that family members of patients with type 1 diabetes tended to have a higher risk of goiter, hypothyroidism, and thyroiditis. Therefore, type 1 diabetes probably shares pathogenetic roots with autoimmune and thyroid diseases.
We did not find a significant familial coaggregation of type 1 diabetes with epilepsy, even though the risk of developing epilepsy among patients with type 1 diabetes has been shown to be ~ 3 times that of the general population both in Taiwan14 and Europe.15 Moreover, a four- to five-fold increased risk of epilepsy has been associated with autoimmune diseases in general, and particularly in children.34 The fact that we failed to detect any familial coaggregation between type 1 diabetes and epilepsy, unlike that seen with other autoimmune disorders, suggests that epilepsy may be a consequence of autoimmune diseases. Our familial aggregation data did not support a common etiology, at least not for genetic factors, between these two disorders.
There are several limitations to the present study. First, the case definition of type 1 diabetes was based purely on diagnoses made by physicians. Nevertheless, the accuracy of the diagnosis of diabetes in the NHI database has previously been validated,20 with one previous study reporting a positive predictive value of type 1 diabetes in the NHI database of 98.3%.6 Therefore, our case definitions are stringent. Second, zygosity of twins is not recorded in the database, so we could not estimate heritability using a classic twin study design. Instead, we used a population-based design to assess the extent of familial aggregation in first-degree relatives. Furthermore, we conducted a validation study which confirmed the accuracy of our method to reconstruct family structure. Third, because we estimated heritability using the threshold liability model which assumes that underlying liability is normally distributed in the population, the results are subject to this assumption. Fourth, assortative mating was not considered in our model. If this assortment was not negligible, heritability could have been underestimated.35 Fifth, this study was restricted to Taiwan, and different findings may occur in different populations and different environments. Therefore, further studies in other countries are required to determine the generalizability of our findings.
Conclusion
This nationwide family study suggests that, in Taiwan, a family history of type 1 diabetes is one of the strongest risk factors for this disease. The differential risk associated with different kinships suggests a strong genetic component to the susceptibility to type 1 diabetes. A family history of type 1 diabetes also exerted an increased risk of other autoimmune and thyroid diseases. This may be useful information when counseling families with one or more affected relatives.
Acknowledgments
This study is based in part on National Health Insurance Research Database data provided by the Applied Health Research Data Integration Service from National Health Insurance Administration. The interpretation and conclusions contained herein do not represent positions of the Administration of National Health Insurance. We would like to thank the Center for Big Data Analytics and Statistics in Chang Gung Memorial Hospital for infrastructure support. This work was funded by the National Science Council of Taiwan (project 105-2314-B-182A-135-MY2) and Chang Gung Memorial Hospital (projects CMRPG3F0833, CORPG3G0231, CMRPG3F0843, CMRPG3G0201, CORPG3F0421, and CORPG3E0143) and was supported by the University of Nottingham for methodology and infrastructure.
Disclosure
The authors report no conflicts of interest in this work.
References
Pugliese A. Advances in the etiology and mechanisms of type 1 diabetes. Discov Med. 2014;18(98):141–150. | ||
Rønningen KS. Environmental trigger(s) of type 1 diabetes: why so difficult to identify? Biomed Res Int. 2015;2015:321656. | ||
Onkamo P, Väänänen S, Karvonen M, Tuomilehto J. Worldwide increase in incidence of Type I diabetes – the analysis of the data on published incidence trends. Diabetologia. 1999;42(12):1395–1403. | ||
Stanescu DE, Lord K, Lipman TH. The epidemiology of type 1 diabetes in children. Endocrinol Metab Clin North Am. 2012;41(4):679–694. | ||
Lu CL, Shen HN, Chen HF, Li CY. Epidemiology of childhood Type 1 diabetes in Taiwan, 2003 to 2008. Diabet Med. 2014;31(6):666–673. | ||
Lin WH, Wang MC, Wang WM, et al. Incidence of and mortality from type I diabetes in Taiwan from 1999 through 2010: a nationwide cohort study. PLoS One. 2014;9(1):e86172. | ||
Parkkola A, Härkönen T, Ryhänen SJ, et al. Extended family history of type 1 diabetes and phenotype and genotype of newly diagnosed children. Diabetes Care. 2013;36(2):348–354. | ||
Noble JA. Immunogenetics of type 1 diabetes: a comprehensive review. J Autoimmun. 2015;64:101–112. | ||
Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–1092. | ||
Patterson CC, Gyürüs E, Rosenbauer J, et al. Trends in childhood type 1 diabetes incidence in Europe during 1989-2008: evidence of non-uniformity over time in rates of increase. Diabetologia. 2012;55(8):2142–2147. | ||
Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52(4):1052–1055. | ||
Kyvik KO, Green A, Beck-Nielsen H. Concordance rates of insulin dependent diabetes mellitus: a population based study of young Danish twins. BMJ. 1995;311(7010):913–917. | ||
Triolo TM, Armstrong TK, Mcfann K, et al. Additional autoimmune disease found in 33% of patients at type 1 diabetes onset. Diabetes Care. 2011;34(5):1211–1213. | ||
Chou IC, Wang CH, Lin WD, et al. Risk of epilepsy in type 1 diabetes mellitus: a population-based cohort study. Diabetologia. 2016;59(6):1196–1203. | ||
Dafoulas GE, Toulis KA, Mccorry D, et al. Type 1 diabetes mellitus and risk of incident epilepsy: a population-based, open-cohort study. Diabetologia. 2017;60(2):258–261. | ||
National Health Insurance Administration DoH, Executive Yuan. The National Health Insurance Statistics, 2015. Available from: http://www.nhi.gov.tw/english/Content_List.aspx?n=70805F6752EE7B9E&topn=616B97F8DF2C3614. Accessed June 27, 2017. | ||
Kuo CF, Grainge MJ, Valdes AM, et al. Familial risk of Sjögren’s syndrome and co-aggregation of autoimmune diseases in affected families: a nationwide population study. Arthritis Rheumatol. 2015;67(7):1904–1912. | ||
Kuo CF, Grainge MJ, See LC, et al. Familial aggregation of gout and relative genetic and environmental contributions: a nationwide population study in Taiwan. Ann Rheum Dis. 2015;74(2):369–374. | ||
Kuo CF, Grainge MJ, Valdes AM, et al. Familial aggregation of systemic lupus erythematosus and coaggregation of autoimmune diseases in affected families. JAMA Intern Med. 2015;175(9):1518–1526. | ||
Lin CC, Lai MS, Syu CY, Chang SC, Tseng FY. Accuracy of diabetes diagnosis in health insurance claims data in Taiwan. J Formos Med Assoc. 2005;104(3):157–163. | ||
Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet. 1990;46(2):222–228. | ||
Lee J, Chia KS. Estimation of prevalence rate ratios for cross sectional data: an example in occupational epidemiology. Br J Ind Med. 1993;50(9):861–862. | ||
Skov T, Deddens J, Petersen MR, Endahl L. Prevalence proportion ratios: estimation and hypothesis testing. Int J Epidemiol. 1998;27(1):91–95. | ||
Barros AJ, Hirakata VN. Alternatives for logistic regression in cross-sectional studies: an empirical comparison of models that directly estimate the prevalence ratio. BMC Med Res Methodol. 2003;3:21. | ||
Biswas A. Statistical Advances in the Biomedical Sciences: Clinical Trials, Epidemiology, Survival Analysis, and Bioinformatics. Wiley series in probability and statistics. Hoboken, NJ: Wiley-Interscience; 2008: 582. | ||
Lin DY. Cox regression analysis of multivariate failure time data: the marginal approach. Stat Med. 1994;13(21):2233–2247. | ||
Whittemore AS, Halpern J. Logistic regression of family data from retrospective study designs. Genet Epidemiol. 2003;25(3):177–189. | ||
Lichtenstein P, Yip BH, Björk C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–239. | ||
Yang J, Visscher PM, Wray NR. Sporadic cases are the norm for complex disease. Eur J Hum Genet. 2010;18(9):1039–1043. | ||
Falconer DS. The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann Hum Genet. 1967;31(1):1–20. | ||
Reich T, James JW, Morris CA. The use of multiple thresholds in determining the mode of transmission of semi-continuous traits. Ann Hum Genet. 1972;36(2):163–184. | ||
Reich T, Rice J, Cloninger CR, Wette R, James J. The use of multiple thresholds and segregation analysis in analyzing the phenotypic heterogeneity of multifactorial traits. Ann Hum Genet. 1979;42(3):371–390. | ||
Hemminki K, Li X, Sundquist J, Sundquist K. Familial association between type 1 diabetes and other autoimmune and related diseases. Diabetologia. 2009;52(9):1820–1828. | ||
Ong MS, Kohane IS, Cai T, Gorman MP, Mandl KD. Population-level evidence for an autoimmune etiology of epilepsy. JAMA Neurol. 2014;71(5):569–574. | ||
Rice TK. Familial resemblance and heritability. Adv Genet. 2008;60:35–49. |
Supplementary material
  | Table S1 ICD-9 codes of diseases in first-degree relatives of patients with type 1 diabetes |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
