Back to Journals » OncoTargets and Therapy » Volume 12
FAM163A, a positive regulator of ERK signaling pathway, interacts with 14-3-3β and promotes cell proliferation in squamous cell lung carcinoma
Authors Liu N, Zhou H, Zhang X , Cai L, Li J, Zhao J, Liu Y , Wang L , Fan C, Li A, Miao Y
Received 7 May 2019
Accepted for publication 23 July 2019
Published 13 August 2019 Volume 2019:12 Pages 6393—6406
DOI https://doi.org/10.2147/OTT.S214731
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Nan Liu,1,* Haijing Zhou,1,* Xiupeng Zhang,1 Lin Cai,1 Jingduo Li,1 Jing Zhao,1 Yang Liu,1 Liang Wang,1 Chuifeng Fan,1 Ailin Li,2 Yuan Miao1
1Department of Pathology, The First Affiliated Hospital and the College of Basic Medical Sciences of China Medical University, Shenyang, People’s Republic of China; 2Department of Radiotherapy, The First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China
*These authors contributed equally to this work
Purpose: FAM163A, also called neuroblastoma-derived secretory protein (NDSP) or C1ORF76, was newly found on chromosome 1q25.2. Previous studies of FAM163A focused on its expression and function in neuroblastoma. However, using an online database, we found that FAM163A may predict poor prognosis in lung squamous cell carcinomas (LUSC). Therefore, the role of FAM163A plays in LUSC needs to be further clarified.
Patients and methods: Western blots, immunofluorescence and immunohistochemistry were used to detect the effect of FAM163A on mediating cell proliferation in vitro and in vivo. Co-immunoprecipitation and immunofluorescence were utilized to evaluate the interaction and co-localization of FAM163A with 14-3-3β and ERK.
Results: In this study, our data revealed that FAM163A overexpression increased the levels of ERK and p90RSK phosphorylation and promoted the expression of cyclin D1. Incorporation with U0126 reversed the effects of FAM163A overexpression. FAM163A directly interacted with both 14-3-3β and ERK and regulated the phosphorylation of ERK by upregulating the protein level of 14-3-3β. Immunohistochemistry results also showed that FAM163A expression significantly correlated with larger tumor size (P=0.023), TNM staging (P=0.015) and regional lymph node metastasis (P=0.016). Kaplan-Meier survival analysis implied the mean survival time of patients with positive FAM163A expression (49.72±3.97 months) was much shorter than the patients with negative FAM163A expression (63.36±3.14 months, P=0.011).
Conclusion: In summary, the present study identified a novel mechanism that FAM163A, through binding and upregulating 14-3-3β, facilitated ERK phosphorylation that led to an increase of cellular proliferation of LUSC cells. FAM163A may be a useful marker to predict poor prognosis of patients with LUSC.
Keywords: FAM163A, ERK, 14-3-3β, prognosis, lung squamous cell carcinomas
Introduction
FAM163A, also called neuroblastoma-derived secretory protein (NDSP) or C1ORF76, is located on chromosome 1q25.2 and encodes a 167 amino acid protein with a putative signal peptide.1,2 Previous studies described that FAM163A specifically overexpressed in neuroblastoma at a higher level than other human cancers. Inhibition of FAM163A expression by RNAi resulted in decreased cellular proliferation and colony formation and led to a downregulation of the levels of ERK phosphorylation.1,2 In another study, they used robust affinity purification-mass spectrometry methodology to elucidate protein interaction networks and predicted that FAM163A may directly binding with multiple 14-3-3 family members, including 14-3-3ζ, 14-3-3γ, 14-3-3ε, 14-3-3β.3 14-3-3 proteins have been proven to express in all eukaryotic cells and are highly conserved in protein sequence and function from yeast to mammals. One of the important functions of 14-3-3 family members is to activate the MAPK-ERK signaling pathway and plays an oncogenic role in cancer development.4–7 Therefore, we speculated that FAM163A might activate ERK signaling pathway through its interaction with 14-3-3 family members.
Lung cancer is the leading cause of cancer-related deaths worldwide.8 Although existing literature about FAM163A was predominantly in neuroblastoma, the expression of FAM163A had been evaluated in several lung cancer cell lines which only presented only negative or weak expression.1,2 The Human Protein Atlas (http://proteinatlas.org/) is an online database which contains immunohistochemical data on most human proteins in a variety of tissues and predicts their prognostic values using TCGA (The Cancer Genome Atlas) RNA samples. In the present study, the expression for FAM163A was analyzed in non-small cell lung cancer (NSCLC) by the above online database. FAM163A was universal expressed in most of the NSCLC tissues. However, overexpression of FAM163A was found predicting poor prognosis in squamous cell lung carcinomas (LUSC) but not in lung adenocarcinomas (Figure S1). To our knowledge, the published literature of FAM163A is limited until now. These observations suggested that FAM163A might play an oncogenic role in LUSC.
In this study, utilizing LUSC cell lines as a model system, we investigated the underlying mechanisms of FAM163A-mediated cellular proliferation in LUSC development. We identified a novel mechanism that FAM163A, through binding and upregulating 14-3-3β, facilitated ERK phosphorylation that led to the increase of cellular proliferation of LUSC cells. Moreover, we also revealed that positive FAM163A expression significantly correlated with poor prognosis in patients with LUSC.
Materials and methods
Cell culture and transfection
The HBE cell line was obtained from ATCC (American Type Culture Collection; Manassas, VA, USA). The SK-MES-1 cell line was obtained from the Shanghai Cell Bank (Shanghai, China). The LK2 cell line was a kindly gift from Prof. Hiroshi Kijima (Department of Pathology and Bioscience, Hirosaki University Graduate School of Medicine, Hirosaki, Japan). All cell lines were authenticated by short tandem repeat (STR) DNA profiling (Figure S2). HBE and LK2 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen), 100 IU/mL penicillin and 100 μg/mL streptomycin (Sigma, St. Louis, MO, USA), SK-MES-1 cell was cultured in MEM medium supplemented with 10% FBS (Invitrogen), 100 IU/mL penicillin and 100 μg/mL streptomycin (Sigma).
Plasmids pCMV6-ddk-myc and pCMV6-ddk-myc-FAM163A (RC203658) were purchased from Origene (Rockville, MD, USA). FAM163A-siRNA (sc-88530) was purchased from Santa Cruz Biotechnology. 14-3-3β-siRNA (SR305137) and control NC-siRNA (SR30004) were purchased from Origene. Stable or transient transfection was carried out using the Lipofectamine 3000 reagent (Invitrogen) as previously described.9 Before we stably transfected FAM163A in LK2 and SK-MES-1 cells, we drew a killing curve of G418 and found that the optimal concentration of G418 was about 100 μg/mL, then we chose 100 μg/mL to select stable transfection cell. By day 14, neo-resistant colonies appeared. Single transgenic colonies were isolated by a micropipette, dissociated into small clumps of cells and transferred into 4-well plates coated with matrigel.The cells proliferated continuously in the presence of G418 and formed a large number of expanding undifferentiated colonies.
Western blotting analysis
Total protein was extracted using lysis buffer (Pierce, Rockford, IL, USA) and quantified using the Bradford method.10 Fifty micrograms of the total protein samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (PVDF; Millipore, Billerica, MA, USA). After blocking with 5% bovine serum albumin (BSA) for 2 hrs at room temperature, the membranes were incubated with the following primary antibodies overnight at 4°C: anti-FAM163A (1:100, sc-390936, Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-GAPDH (WH0002597M1, 1:5000, Sigma); anti-Myc-tag (#2276), anti- cyclin B1 (#4138), anti-cyclin D1 (#2978), anti-cyclin E1 (#4129), anti-cyclin H (#2927), anti-phosphorylated(p)-P38 ((Thr180/Tyr182#4511), P38 (#2308), anti-active-β-catenin (Ser45, #19807), anti-β-catenin (#2698), anti-p-JNK (Thr183/Tyr185, #4668), anti-JNK(#9252), anti-p-NF-κB (Ser536#13346), anti-NF-κB (#8242), anti-p-AKT (Ser473#12694), anti-AKT (#2920), anti-p-ERK (Thr202/Tyr204, #4370), anti-ERK (#4695) anti-p-p90RSK (Ser380, #11989), p90RSK (#9355), anti-p-GSK3β (Ser9, #5558), anti-GSK3β(#12456), anti-14-3-3β (#9636), anti-14-3-3γ (#5522), anti-14-3-3ε (#9635), 14-3-3ζ/δ (#7413), (1:500; Cell Signaling Technology, Danvers, MA, USA). After washing, membranes were incubated with peroxidase-conjugated IgG (anti-mouse or anti-rabbit, Cell Signaling Technology) at 37°C for 2 hrs. Bound proteins were visualized by ECL (electrochemiluminescence, Pierce) and detected with a bio-imaging system (DNR Bio-Imaging Systems, Jerusalem, Israel).
Immunofluorescent staining
The HBE, LK2 and SK-MES-1 cells were fixed with 4% paraformaldehyde, blocked with 1% bovine serum albumin (BSA) and then incubated with the FAM163A (1:50, sc-390936, Santa Cruz Biotechnology), anti-ERK (#4695, Cell Signaling Technology) and anti-14-3-3β (#9636, Cell Signaling Technology) overnight at 4°C, followed by incubation with tetramethylrhodamine isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (1:50, Zhongshan Golden Bridge, Beijing, China) at 37°C. The nuclei were counterstained with DAPI (4′, 6-diamidino-2-phenylindole). Epifluorescent microscopy was performed using a Radiance 2000 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY, USA).
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazoliumbromide (MTT) assay
Cells were plated in 96-well plates with medium containing 10% FBS in a density of 3,000 cells per well. Five days later, 10 μL of 5 mg/mL MTT solution was added into each well and incubated for 4 hrs at 37°C, the medium was then removed from each well, and the resultant MTT formazan was solubilized in 150 μL of DMSO (dimethyl sulfoxide). The results were quantified spectrophotometrically at a wavelength of 490 nm. Each test carried out in triplicate.
Colony formation assay
LK2 and SK-MES-1 cells were stably transfected with pCMV6 or pCMV6-FAM163A plasmids, pretreated with U0126 or 14-3-3β siRNA. After that, cells were plated into three 6-cm cell culture dishes (1,500 cells per dish for LK2 and SK-MES-1 cell lines) and incubated for 12 days. Plates were washed with phosphate-buffered saline (PBS) and stained with Hematoxylin. The number of colonies with more than 50 cells was counted. Each experiment was carried out in triplicate.
Xenograft model
The animals used in this study were treated according to the National Institutes of Health Guide for the care and use of Laboratory Animals (NIH Publications No. 8023, revised1978). Four-week-old female BALB/c nude mice were purchased from SLAC Laboratory Animal (Shanghai, China) and raised in a laminar flow cabinet under specific pathogen-free conditions for two weeks before use. Mice were randomized into different groups (n=5 per group). LK2 cells stably transfected with either empty plasmid or FAM163A plasmid were injected subcutaneously into the nude mice. Bidimensional tumor measurements were taken on the 12th day and the 36th day; after which the mice were sacrificed. All experimental procedures were approved by the Animal Ethics Committee of China Medical University. Each experiment was carried out in triplicate.
Co-immunoprecipitation (Co-Ip)
LK2 and SK-MES-1 cells transfected with FAM163A plasmid were harvested with 1X Lysis/Wash Buffer plus PMSF, and incubated with Protein A/G-agarose (Santa Cruz Biotechnology) at 4°C overnight. After that, the lysates were immunoprecipitated overnight at 4°C with either anti-FAM163A (1:50, sc-390936, Santa Cruz Biotechnology) or anti-mouse IgG antibody bound to protein G PLUS-agarose (Santa Cruz Biotechnology). The immunoprecipitated proteins were evaluated by Western blotting assay. Each experiment was carried out in triplicate.
Patients and specimens
This study was conducted with the approval of the local institutional review board at China Medical University (LS2016 [009]). Written consent was signed by each participant for their specimens to be used in this study. Written informed consent was given by the participants for their information to be stored in the hospital database for their specimens to be used in this study. All clinical investigation has been conducted according to the principles expressed in the Declaration of Helsinki. Tissue samples were obtained from 83 patients (58 males and 25 females) who were received surgical excision at the First Affiliated Hospital of China Medical University between 2010 and 2012, with a diagnosis of LUSC. The mean age of the 83 patients was 60 years old (ranging from 29 to 79 years old). Histological diagnosis and grading were evaluated according to the 2015 World Health Organization classification of tumors of the lung.11 Tumor staging was performed according to the seventh edition of the Union for International Cancer Control TNM Staging System for Lung Cancer.12 No neoadjuvant radiotherapy or chemotherapy was administered before surgery. The overall survival was defined as the time from the day of surgery to the end of follow-up or the day of death. Sixteen freshly isolated specimens, including both tumor tissue and the corresponding normal tissues, were stored at −80°C immediately after resection for protein extraction.
Real-time PCR
Total RNA was extracted from 14 pairs of lung squamous cell carcinoma tissues and corresponding normal tissues using an RNeasy RNA isolation kit (QIAGEN, Hilden, Germany). First-strand cDNA was synthesized from 1 μg of total RNA using ReverTra Ace (TOYOBO, Osaka, Japan). RT-PCR was performed using an aliquot of first-strand cDNA as a template under standard conditions with Taq DNA polymerase (QIAGEN). The real-time polymerase chain reaction (PCR) was carried out using SYBR Green Assay (Applied Biosystems, Tokyo, Japan). For the SYBR Green system, the following forward and reverse primers were used for NDSP and β-actin (internal control): FAM163A, forward, GCTACGGTGATCCTCCTCTG, and reverse, CTCCTC CTCGTCTGCAACCT; β-actin, forward, AGAGAGGCATCCTCACCCTG, and reverse, CATGAGGTAGTCAGTCAGGT.1
Immunohistochemistry
The samples sections were incubated with anti-FAM163A (1:200, sc-390936, Santa Cruz Biotechnology). Immunohistochemistry staining was performed and scored as previously described.9 A final score of 0–12 was obtained by multiplying the intensity by the percentage scores. Tumors were regarded as positive for FAM163A expression if their score was>4. Tumor samples with scores between 1 and 4 were categorized as showing weak expression, whereas those with scores of 0 were considered to have no expression; both weak expression and no expression were defined as negative FAM163A expression.
Statistical analysis
SPSS version 22.0 for Windows (SPSS, Chicago, IL, USA) was used for all statistical analyses. Pearson’s Chi-square test was applied to evaluate possible correlations between FAM163A and clinicopathological factors. The Mann–Whitney U test was used for the image analysis of colony formation assay, MTT, xenograft assay. Kaplan–Meier survival analysis was carried out for the 83 LUSC cases and compared using the log-rank test. A P<0.05 was regarded as statistically significant.
Results
Overexpression of FAM163A promoted cell proliferation in lung squamous cell carcinoma in vivo and in vitro
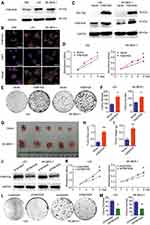
We initially evaluated the protein levels of FAM163A in normal bronchial epithelial cell line (HBE) and two LUSC cell lines LK2 and SK-MES-1 using Western blotting. FAM163A was observed in all three cell lines, and the protein level in LK2 and SK-MES-1 was higher than that in HBE (Figure 1A). As shown in Figure 1B, immunofluorescence assay suggested that FAM163A was positively expressed in the cytoplasm of HBE, LK2 and SK-MES-1 cell lines. To investigate the effect of FAM163A on proliferation, we overexpressed FAM163A in LK2 and SK-MES-1cells, the efficiency of transfection was shown in Figure 1C. Subsequent cell viability assays indicated that cell transfected with FAM163A plasmid grew more rapidly than those transfected with the empty vector in LK2 and SK-MES-1 cell lines (P<0.05, respectively; Figure 1D). Colony formation assays results presented similar tendency compared with the controls (P<0.05, respectively; Figure 1E and F). In agreement with the results from in vitro analyses, our in vivo experiments revealed that the size and weight of the tumor xenografts were dramatically increased in mice injected with cells overexpressing FAM163A compared with those injected with cells transfected with empty vector (P<0.01, respectively; Figure 1G–I). Then we interfered the expression of FAM163A in LK2 and SK-MES-1 cells, the efficiency was shown in Figure 1J. Subsequent cell viability assays (Figure 1K) and colony formation assays (Figure 1L and M) indicated that FAM163A RNAi inhibited the proliferation in both LK2 and SK-MES-1 cell lines (P<0.05, respectively).
FAM163A upregulated the expression of cyclin D1 via phosphorylation of the ERK/p90RSK signaling pathway
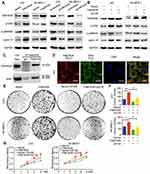
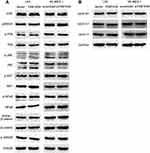
To explore the mechanism of FAM163A promoting cell proliferation, we initially evaluated various key pathways proteins and its downstream target proteins related to cell growth. The results of Western blotting suggested that the expression of p-ERK, p-p90RSK and cyclin D1 were upregulated after FAM163A overexpression and downregulated by interfering FAM163A expression (Figure 2A). However, the expression of ERK, p90RSK, p-P38, P38, p-JNK, JNK, p-AKT, AKT, p-NFκB, NFκB, Active-β-catenin, β-catenin, p-GSK3β, GSK3β, cyclin B1, cyclin E1, and cyclin H did not show visible changes compared with the controls (Figure S3). By treated LK2 and SK-MES-1 cells with MEK inhibitor U0126, we found that the effects of FAM163A upregulating the phosphorylation of ERK and p90RSK, as well as the protein level of cyclin D1 were attenuated by U0126 incorporation (Figure 2B). For further investigating the relationship between FAM163A and ERK signaling pathway, we performed co-IP assays. The results indicated FAM163A directly interacted with ERK (Figure 2C). The following immunofluorescence also found that FAM163A and ERK co-localized in the cytoplasm of LK2 cells (Figure 2D). Besides, subsequent colony formation assays (Figure 2E and F) and MTT assays (Figure 2G) also revealed that the increase of cellular proliferation caused by FAM163A cDNA transfection was significantly reversed by U0126 incorporation.
FAM163A promoted the phosphorylation of the ERK/p90RSK signaling pathway by interacting with 14-3-3β
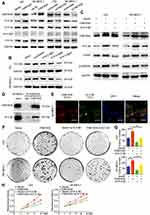
We first examined the expression of 14-3-3 family proteins after overexpressing FAM163A and interfering the expression of FAM163A in LK2 and SK-MES-1 cells. The Western blots results revealed that the expression of 14-3-3β was increased along with FAM163A overexpression and downregulated when FAM163A knockout. While the expression of 14-3-3γ, 14-3-3ζ/δ and 14-3-3ε showed no visible changes compared with the controls (Figure 3A). Next, we used three small interfering RNA to knock down 14-3-3β in both LK2 and SK-MES-1 cells, si14-3-3β#3 was applied to perform the subsequent experiments (Figure 3B). We found the upregulation of ERK and p90RSK phosphorylation resulted from overexpressing FAM163A was reversed by 14-3-3β RNAi (Figure 3C). The subsequent co-IP assays suggested that FAM163A interacted with 14-3-3β (Figure 3D). The confocal microscopic analysis further confirmed this interaction (Figure 3E). Moreover, we examined whether 14-3-3β inhibition affected the functional role of cells using colony formation assays. We found 14-3-3β RNAi significantly reduced the number of viable colonies increased by overexpression of FAM163A (P<0.05, Figure 3F and G), as well as cell growth (Figure 3H).
FAM163A positively expressed in the cytoplasm and predicted
Poor clinical outcomes of patients with lung squamous cell
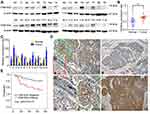
Western blotting analysis showed that FAM163A protein level in 16 pairs of lung squamous cell carcinoma tissues and their corresponding normal tissues. Eleven of sixteen (68.75%) samples of lung squamous cell carcinoma showed higher expression compared with their corresponding noncancerous tissues (Figure 4A). The mean protein level of FAM163A in lung squamous cell carcinoma tissues (mean ± SE, 0.76±0.05) was significantly higher than that in the corresponding normal tissue (mean ± SE, 0.51±0.07, P=0.0267, Figure 4B). Next, the mRNA level of FAM163A in 14 pairs of lung squamous cell carcinoma tissues and their corresponding normal tissues was tested using PCR. Of the 14 patients, 10 (71.4%) showed a higher expression level of FAM163A mRNA in lung cancer tissue specimens than in noncancerous tissue specimens (Figure 4C). The mean expression value of FAM163A mRNA in cancer tissues (43.61±25.45; mean ± SD, normalized by beta-actin gene expression) was significantly higher than the value (26.18±15.14) in the corresponding noncancerous tissues (P<0.05). Using immunohistochemistry, we next examined FAM163A expression in 83 lung squamous cell carcinoma tissues. Among the 83 patients, 32 were 60 years old or older and 51 patients were less than 60 years old. A total of 74 tumors were well or moderately differentiated, while 9 were classified as poorly differentiated. Lymph node metastases were present in 33 of the 83 cases. The tumors included 63 cases at stages I–II and 20 cases at stage III. We found FAM163A expression was strongly in the cytoplasm of 47/83 (56.6%) samples (Figure 4D). Statistical analysis results suggested that FAM163A expression significantly correlated with larger tumor size (P=0.023), TNM staging (P=0.015) and regional lymph node metastasis (P=0.016). FAM163A expression showed no significant correlation with age, gender and histological differentiation (P>0.05, Table 1). Kaplan-Meier survival analysis implied the mean survival time of patients with positive FAM163A expression (49.72±3.97 months) was much shorter than the patients with negative FAM163A expression (63.36±3.14 months, P=0.011, Figure 4E).
 |
Table 1 The association between FAM163A expression and the clinicopathological characteristics |
Discussion
In the present study, we identified that FAM163A interacted with 14-3-3β and ERK, which thereby facilitating the phosphorylation of ERK by upregulating the protein level of 14-3-3β. Subsequently, FAM163A increased the levels of p90RSK phosphorylation and promoted the expression of cyclin D1, which were downstream molecules of ERK signaling, and upregulated proliferation of LUSC cells. In clinical tissue samples, positive FAM163A expression significantly correlated with larger tumor sizes, advanced TNM staging, positive regional lymph node metastasis and poor overall survival.
To our knowledge, the publishing literature refers to FAM163A is limited. The opinion of previous studies considered that FAM163A was specifically overexpressed in neuroblastoma at higher levels than other tissues including malignancies and normal tissues.2 In the same study, they also revealed that FAM163A was a secreted protein in several neuroblastoma cell lines.2 However, we had evaluated the supernatant protein of LUSC cells with or without FAM163A cDNA transfection and found no visible secreted FAM163A (data not shown). Thus, we hypothesized that FAM163A might also play a role in the cytoplasm in LUSC cells. In the present study, using an online database (http://proteinatlas.org/), we found that FAM163A also expressed in LUSC tissues. Our results of immunoperoxidase staining revealed that FAM163A positively expressed both in the cytoplasm of LUSC cells and LUSC tissues, as well as their non-cancerous counterparts. Since FAM163A has been identified to be an oncogene in neuroblastoma, the expression of FAM163A in LUSC makes one wonder if it also plays an oncogenic role in the development of LUSC.1,2 Using FAM163A cDNA transfection, we found that FAM163A promoted cell proliferation of LUSC both in vitro and in vivo. Our data were consistent with previous studies that FAM163A facilitated cell proliferation in neuroblastoma.2 In the same study, they addressed that FAM163A induced cell proliferation was mediated by ERK activation.2 To confirm that, we evaluated not only ERK signaling but also the other signaling which was considered to be related to cell proliferation, such as P38, JNK, AKT, NFκB and WNT signaling. Our results also showed that overexpression of FAM163A upregulated the phosphorylation of ERK, as well as its downstream molecule (p90RSK). The previous studies and ours indicated that FAM163A-ERK axis might play a role in regulating cell proliferation of malignancies.1,2 We then proceeded to assess the effect of FAM163A overexpression on cell cycle genes and revealed that cyclin D1 expression was upregulated. As cyclin D1 was a downstream molecule of ERK signaling, we next incorporated ERK inhibitor to observe whether FAM163A promote cyclin D1 expression through ERK signaling.13,14 Our results showed that FAM163A promoted cell proliferation through interacting and activating ERK-cyclin D1 signaling. Although previous studies have demonstrated that FAM163A regulated ERK signaling pathway, the results of the present study revealed that FAM163A directly interacted with ERK.2
The 14-3-3 family member involved in various cellular processes including signal transduction, cell cycle, cell proliferation, apoptosis, differentiation and survival.15–17 Furthermore, 14-3-3 family members are able to activate the MAPK-ERK signaling pathway and plays an oncogenic role in cancer development.4–7 In a previous study, using affinity purification-mass spectrometry, they predicted that FAM163A might directly bind with 14-3-3 family members.3 Although the mechanisms of the binding between FAM163A and 14-3-3 family are unclear, our results showed that the protein level of 14-3-3β was significantly elevated after overexpression of FAM163A. Subsequent co-IP results suggested that FAM163A directly interacted with 14-3-3β. As we had confirmed that FAM163A directly interacted with ERK and ERK signaling was a putative downstream signaling pathway of 14-3-3 family members in regulating the development of human neoplasms, we next inhibited the expression of 14-3-3β in FAM163A overexpression cells.4–7 Western blotting results showed that the increase of the levels of phosphorylated ERK and p90RSK induced by FAM163A overexpression were attenuated by 14-3-3β RNAi, as well as the results of colony formation analysis and MTT analysis. These results suggested that FAM163A, 14-3-3β and ERK were likely to form a complex and FAM163A might facilitate the phosphorylation of ERK signaling through interacting with 14-3-3β which thereby mediated cell proliferation of LUSC cells.
In the previous study, the mRNA levels correlate with risk group assignment in patients with neuroblastoma.1 In our study, we found that FAM163A expression significantly correlated with larger tumor size, TNM staging, regional lymph node metastasis and predicted poor prognosis in patients with LUSC. Our data were consistent with the results from an online database which indicated that FAM163A might have prognostic value in patients with LUSC.
Conclusion
In summary, the present study identified a novel mechanism that FAM163A facilitates ERK phosphorylation through binding and upregulating 14-3-3β, which led to an increase in cellular proliferation of LUSC cells. Moreover, we also revealed that positive FAM163A expression significantly correlated with poor prognosis in patients with LUSC.
Acknowledgments
We thank Dr. Hiroshi Kijima for providing the LK2 cell line. This work was supported by the National Natural Science Foundation of China (No. 81472805 to Yuan Miao, No. 81602012 to Xiupeng Zhang, and No. 81402520 to Ailin Li), the Natural Science Foundation of Liaoning Province to Yuan Miao (No. 201421044) and the Research Foundation for the Doctoral Program to Ailin Li (No. 20141040).
Author contributions
All authors contributed to data analysis, drafting or revisiong the artical, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Vasudevan SA, Russell HV, Okcu MF, et al. Neuroblastoma-derived secretory protein messenger RNA levels correlate with high-risk neuroblastoma. J Pediatr Surg. 2007;42(1):148–152.
2. Vasudevan SA, Shang X, Chang S, et al. Neuroblastoma-derived secretory protein is a novel secreted factor overexpressed in neuroblastoma. Mol Cancer Ther. 2009;8(8):2478–2489.
3. Huttlin EL, Bruckner RJ, Paulo JA, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509.
4. Freeman AK, Morrison DK. 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol. 2011;22(7):681–687.
5. Hirotsune S. [Unique role of LIS1 and NDEL1 as a regulator of cytoplasmic dynein]. Tanpakushitsu Kakusan Koso Protein, Nucleic Acid, Enzyme. 2006;51(6 Suppl):586–590.
6. Sluchanko NN. Association of multiple phosphorylated proteins with the 14-3-3 regulatory hubs: problems and perspectives. J Mol Biol. 2018;430(1):20–26.
7. Khorrami A, Sharif Bagheri M, Tavallaei M, Gharechahi J. The functional significance of 14-3-3 proteins in cancer: focus on lung cancer. Horm Mol Biol Clin Investig. 2017;32(3):1868–1891.
8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
9. Zhou H, Cai L, Zhang X, et al. ARHGEF39 promotes tumor progression via activation of Rac1/P38 MAPK/ATF2 signaling and predicts poor prognosis in non-small cell lung cancer patients. Lab Invest. 2018;98(5):670–681.
10. Kruger NJ. The bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15.
11. Travis WD, Brambilla E, Burke A, Marx A, Nicholson AG. WHO classification of tumors of the lung, pleura, thymus and heart. Lyon Int Agency Res Cancer. 2015.
12. Edge SB, Compton CC. The American joint committee on cancer: the7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474.
13. Schwartz MA, Assoian RK. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114(Pt 14):2553–2560.
14. Assoian RK, Klein EA. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008;18(7):347–352.
15. Aitken A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1996;6(9):341–347.
16. Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16(3):162–172.
17. Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113(3):359–363.
Supplementary materials
 |
Figure S1 The results of online prognostic analysis. (A) The K-M plot of patients with squamous cell lung carcinomas (P=0.037). (B) The K-M plot of patients with adenocarcinomas (P=0.10). |
 |
Figure S2 The certification of STR DNA profiling. (A) The certification of LK2 was provided by JCRB cell bank. (B) The certification of SK-MES-1 was provided by Chinese Academy of Sciences Cell Bank. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.