Back to Journals » OncoTargets and Therapy » Volume 12
EZH2 Contributes To Cisplatin Resistance In Breast Cancer By Epigenetically Suppressing miR-381 Expression
Authors Dou D, Ge X, Wang X, Xu X, Zhang Z, Seng J, Cao Z, Gu Y, Han M
Received 1 May 2019
Accepted for publication 23 September 2019
Published 13 November 2019 Volume 2019:12 Pages 9627—9637
DOI https://doi.org/10.2147/OTT.S214104
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Dongwei Dou,* Xin Ge,* Xinxing Wang, Xiaodong Xu, Zhe Zhang, Jingjing Seng, Zhang Cao, Yuanting Gu, Mingli Han
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 475000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mingli Han
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, 1 Jianshe East Road, Zhengzhou 450000, Henan Province, People’s Republic of China
Tel +86-371-66273586
Email [email protected]
Background: Emerging evidence reveals the vital role of enhancer of zeste homolog 2 (EZH2) in cancer chemoresistance. However, its function and molecular mechanisms in breast cancer chemoresistance remain largely unknown.
Methods: Gene expression was evaluated using quantitative real-time PCR (qRT-PCR) and Western blot analysis. The functional roles of EZH2 and miR-381 in breast cancer were explored using cell MTT assay and flow cytometry analysis. The effect of EZH2 on miR-381 expression in transcriptional level was determined using Chromatin immunoprecipitation (ChIP) assay and Luciferase reporter assay.
Results: In this study, we found that EZH2 was up-regulated in CDDP-resistant breast cancer tissues and cell lines. Breast cancer patients with high EZH2 expression had a poor prognosis. EZH2 silencing improved the sensitivity of MCF-7/CDDP and MDA-MB-231/CDDP cells towards CDDP. Moreover, EZH2 could epigenetically silence miR-381. miR-381 overexpression could overcome CDDP resistance in CDDP-resistant breast cancer cells. miR-381 knockdown weakened the inductive effect of EZH2 silencing on CDDP sensitivity of MCF-7/CDDP and MDA-MB-231/CDDP cells. Furthermore, EZH2 knockdown facilitated CDDP sensitivity of CDDP-resistant breast cancer cells in vivo.
Conclusions: Collectively, EZH2 depletion overcame CDDP resistance of breast cancer through epigenetically silencing miR-381, providing a novel therapeutic target for breast cancer chemoresistance.
Keywords: breast cancer, CDDP, enhancer of zeste homolog 2, miR-381
Introduction
Breast cancer is a most common malignancy in women worldwide and a major serious health threat to women.1 Even though impressive improvements have been acquired in diagnosis and therapy of breast cancer during the past decade, prognosis for advanced breast cancer remains relatively poor.2 Platinum-based chemotherapy is a significant therapeutic strategy for breast cancer patients.3 Nevertheless, chemoresistance frequently occurs during chemotherapy, which is a crucial barrier to the efficacy of chemotherapy drugs for cancers, including breast cancer.4,5 Consequently, to sequentially elucidate the underlying mechanism and discover new therapeutic targets are essential for developing effective therapies for breast cancer patients.
Enhancer of zeste homolog 2 (EZH2), a histone methyltransferase, is a vital catalytic subunit of polycomb repressive complex 2 (PRC2) epigenetically suppressing gene expression through the enhancement of histone H3 lysine 27 trimethylation (H3K27me3).6,7 EZH2 was aberrantly up-regulated in various cancers, and was closely linked to the tumor progression, metastasis and poor prognosis.8–10 Moreover, EZH2 expression was remarkably increased in breast cancer cells and contributed to breast cancer progression.11,12 Despite the involvement of EZH2 in cancer progression extensively characterized, the role of EZH2 in acquired drug resistance remains elusive. Therefore, to investigate whether EZH2 inhibition will hold promise in the treatment of resistant cancers is urgent.
miRNAs are a class of small, non-coding RNA with about 22 nucleotides in length, which contribute to their target genes’ inhibition, through mRNA degradation or translation inhibition.13,14 MiRNAs are recently implicated by more and more reports as critical determinants that are involved in various cellular processes, including chemoresistance and tumorigenesis.15 MiRNAs’ aberrant expression has been regarded as a powerful regulator of CDDP resistance. For instance, miR-34a has been discovered to be down-regulated and contributed to CDDP resistance in CDDP-resistant prostate cancer cells.16 miR-381 was reported to be decreased in breast cancer, and miR-381 overexpression suppressed breast cancer cell proliferation, epithelial-to-mesenchymal transition and metastasis through targeting CXCR4.17 Additionally, miR-381 could re-sensitize CDDP-resistant non-small cell lung cancer (NSCLC) cells towards cisplatin through inactivating nuclear factor-κB signaling.18 Moreover, miR-381 enhanced doxorubicin sensitivity of breast cancer cells through inactivation of MAPK signaling via targeting FYN.19 Recently, miR-381 was reported to overcome CDDP sensitivity in breast cancer through targeting MDR1.20 All these studies suggested that miR-381 was closely implicated with cancer chemoresistance. However, how miR-381 was regulated in breast cancer is still largely unknown.
In this study, we aimed to investigate the expression pattern and functional role of EZH2 in breast cancer CDDP resistance. Our results discovered that EZH2 expression was up-regulated in CDDP-resistant breast cancer tissues and cells. Functionally, EZH2 silencing improved the response of CDDP-resistant breast cancer cells toward CDDP. Mechanically, EZH2 knockdown facilitated the sensitivity of breast cancer cells towards CDDP through epigenetic suppression of miR-381 expression. Our work demonstrated a novel EZH2/miR-381 regulatory axis, overcoming CDDP resistance in breast cancer.
Materials And Methods
Tumor Tissue Samples And Cells
The paired tumor tissues (n=48) and adjacent normal tissues (n=48) were obtained from breast cancer patients who underwent surgery at the First Affiliated Hospital of Zhengzhou University. All patients were treated with CDDP and divided into two groups: CDDP-sensitive (relieved completely or partially) and CDDP-resistant (stable or deteriorating) patients. The informed consent has been signed by all patients before the study, which was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. Normalized RNA-seq data of Breast adenocarcinoma (BRCA) were downloaded from the TCGA data portal website (https://cancergenome.nih.gov/).
Human breast cancer cell lines (MCF-7 and MDA-MB-231) and human normal breast epithelial cell line MCF-10A were derived from ATCC (Manassas, VA, USA). CDDP-resistant cell lines (MCF-7/CDDP and MDA-MB-231/CDDP) were chosen from MCF-7 and MDA-MB-231 cells after continuous exposure to stepwise increasing concentrations of DDP for 12 months. All cells were grown in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% FBS at 37°C with 5% CO2.
Cell Transfection
Mimic control (miR-con), miR-381 mimic (miR-381) and miR-381 inhibitor (anti-miR-381) were purchased from Sangon Biotech (Shanghai, China). The empty pcDNA3.1 vector (Vector) or EZH2 overexpressing vector pcDNA3.1-EZH2 (EZH2) and small interfering RNA against EZH2 (si-EZH2) and its negative control (si-con) were designed and synthesized by Genepharma (China). Lipofectamine 2000 (Invitrogen) was used for cell transfections.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA extraction was carried out through using Trizol reagent (TaKaRa, Tokyo, Japan). Then, the RNA was reverse transcripted to cDNA using PrimeScript RT Reagent Kit (TaKaRa). The qRT-PCR was carried out using SYBR green qRT-PCR assay. GAPDH and U6 were used for internal controls for EZH2 and miR-381, respectively. The data were analyzed using the 2−ΔΔCt method.
Western Blot Analysis
Equivalent amounts of protein samples were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polypropylene difluoride membranes. After 1 hr of blockade with 5% non-fat dry milk, membranes were immunoblotted overnight at 4°C with the special primary antibody against EZH2 (BD Biosciences, San Jose, CA, USA), followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (Boster, Wuhan, China) for 1 hr at room temperature. Visualization of bands was performed according to the ECL kit (Beyotime, Shanghai, China).
CDDP Sensitivity Assay
The sensitivity of breast cancer cells was evaluated by MTT assay. CDDP sensitivity was determined using the IC50 value (half maximal inhibitory concentration). GraphPad Prism 7.0 Software was used to calculate the IC50 value.
Flow Cytometric Analysis
Cell apoptosis was identified using Annexin V-FITC/PI apoptosis detection kit (MultiScience Biotech, Hangzhou, China) as described previously.21
Chromatin Immunoprecipitation (ChiP) Assays
ChIP assay was conducted using EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA) with anti-EZH2 (Abcam, Cambridge, MA, USA), anti-H3K27me3 (Millipore) or anti-IgG (Millipore) antibodies. Precipitated DNA was tested by qPCR.
Luciferase Reporter Assay
The miR-381 promoter reporter vector was purchased from Genechem (China). Then, MCF-7/CDDP cells were co-transfected with (EZH2 or Vector) or (si-EZH2 or si-con) and the reporter promoter. Finally, luciferase activity in MCF-7/CDDP cells was measured using Luciferase Reporter assay system (Promega, Madison, WI, USA).
Animal Experiments
The animal experiments were performed in strict accordance with the guiding principles of the institutional animal ethics committee and approved by the institutional research committee of the First Affiliated Hospital of Zhengzhou University. MCF-7/CDDP cells stably expressing sh-EZH2 were transplanted into six-week-old BALB/c nude mice (5 mice/group) from Slac Laboratory (Shanghai, China), and then mice were injected intraperitoneally with 5 mg/kg CDDP or a same volume of PBS every seven days. Every seven days, tumor sizes were monitored and calculated using the formula: volume = 0.5 × length × width2. Mice were euthanized at 35th day post-injection and tumors were excised and weighed.
Statistical Analysis
All data were evaluated as means ± standard deviation (SD). Student’s t-test and one-way ANOVA were used to calculate the statistic difference. P value <0.05 was considered statistically significant.
Results
EZH2 Was Up-Regulated In CDDP-Resistant Breast Cancer Tissues And Cell Lines
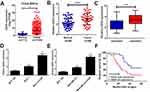
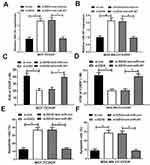
To investigate the role of EZH2 in breast cancer, we firstly studied the expression of EZH2 in breast cancer tissues from TCGA databases. Compared with normal tissues, EZH2 expression was remarkably up-regulated in breast cancer tumor tissues (Figure 1A). To verify the result from TCGA databases, EZH2 expression in breast cancer tumor tissues (n=48) and adjacent normal tissues (n=48) was further determined by qRT-PCR analysis. Consistently, EZH2 expression was higher in breast cancer tissues than that in adjacent normal tissues (Figure 1B). Moreover, compared with CDDP-sensitive breast cancer tissues, EZH2 expression was extremely increased in CDDP-resistant breast cancer tissues (Figure 1C). Furthermore, the expression of EZH2 was significantly enhanced in MCF-7 and MDA-MB-231 cells compared with normal MCF-10A cells (Figure 1D and E). Notably, compared with their parental cells, MCF-7/CDDP and MDA-MB-231/CDDP cells showed high EZH2 expression level (Figure 1D and E). Moreover, the breast cancer patients with high EZH2 level had a poor prognosis (P = 0.0165) (Figure 1F). Collectively, these data seemed to suggest that up-regulated EZH2 may be implicated with CDDP resistance in breast cancer.
EZH2 Knockdown Overcame CDDP Resistance Of Breast Cancer Cells
To evaluate the resistance of MCF-7/CDDP and MDA-MB-231/CDDP cells to CDDP, IC50 of CDDP was measured by MTT assay in CDDP-resistant MCF-7/CDDP and MDA-MB-231/CDDP cells and parental MCF-7 and MDA-MB-231 cells. Compare to MCF-7 and MDA-MB-231 cells, MCF-7/CDDP and MDA-MB-231/CDDP cells displayed poor response to CDDP (Figure 2A). To further confirm the role of EZH2 in CDDP-resistant breast cancer cells, MCF-7/CDDP and MDA-MB-231/CDDP cells were transfected with EZH2 siRNAs (si-EZH2 #1, si-EZH2 #2 or si-EZH2 #3) or si-con. qRT-PCR analysis indicated that the introduction of EZH2 siRNAs evidently weakened EZH2 expression in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 2B), especially in si-EZH2 #2 treated group. Therefore, si-EZH2 #2 (si-EZH2) was employed to further experiments. Dramatically, EZH2-silencing suppressed the cell viability and enhanced CDDP sensitivity in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 2C and D). To further determine the role of EZH2 in CDDP-induced apoptosis, flow cytometry analysis was undertaken in MCF-7/CDDP and MDA-MB-231/CDDP cells with 10 μM CDDP exposure. As expected, EZH2 knockdown strangely enhanced CDDP-induced apoptosis in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 2E and F). Collectively, EZH2 knockdown facilitated CDDP sensitivity in breast cancer cells.
EZH2 Epigenetically Suppressed miR-381 Expression In Breast Cancer Cells
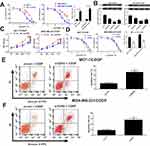
Previous studies demonstrated that EZH2 could contribute to transcriptional inhibition of miRNAs through increasing H3K27me3 on their promoter region.22,23 Moreover, Chipbase database (http://rna.sysu.edu.cn/chipbase/) prediction indicated that EZH2 could bind with miR-381, which was identified to be down-regulated in cancers attributed to DNA hypermethylation.24 Hence, whether EZH2 contributed to epigenetic suppression of miR-381 in CDDP-resistant breast cancer cells was further investigated. Firstly, the correlation between EZH2 and miR-381 in 1185 breast cancer tissue samples from TCGA databases was analyzed using Chipbase database. The results indicated that there was a negative correction between EZH2 and miR-381 expression in breast cancer tissue samples (Figure 3A). EZH2 knockdown evidently increased miR-381 expression in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 3B). Moreover, EZH2-silencing considerably elevated precursor miR-381 expression in MCF-7/CDDP and MDA-MB-231/CDDP cells (Supplement Figure 1A and B). To further confirm that miR-381 is transcriptionally repressed by EZH2 in our breast cancer cells, ChIP assays were conducted to detect the enrichment of EZH2 and the H3K27me3 on the miR-381 promoter. The results disclosed that EZH2 knockdown remarkedly weakened the enrichment of EZH2 and H3K27me3 on the miR-381 promoter in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 3C and D). Also, luciferase reporter assay showed that EZH2 inhibition improved the activity of miR-381 promoter, oppositely, up-regulation of EZH2 suppressed the promoter activity (Figure 3E and F). All these data suggested that EZH2 epigenetically suppressed miR-381 expression in breast cancer cells.
miR-381 Overexpression Enhanced CDDP Sensitivity Of Breast Cancer Cells
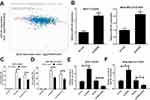
To further study the effect of miR-381 on CDDP-resistant breast cancer cells, MCF-7/CDDP and MDA-MB-231/CDDP cells were transfected with miR-381 mimics or miR-con. qRT-PCR analysis revealed that miR-381 expression was remarkably increased in miR-381 transfecting MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 4A and B). Moreover, miR-381 overexpression improved the sensitivity of MCF-7/CDDP and MDA-MB-231/CDDP cells to CDDP (Figure 4C and D). To further determine the effect of miR-381 on CDDP-induced apoptosis, flow cytometry analysis was performed in MCF-7/CDDP and MDA-MB-231/CDDP cells exposed to 10 μM CDDP. As expected, miR-381 overexpression strangely enhanced CDDP-induced apoptosis in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 4E and F). Together, elevated EZH2 improved CDDP sensitivity in breast cancer cells.
EZH2 knockdown facilitated CDDP sensitivity of breast cancer cells through increasing miR-381 expression
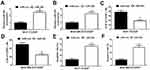
To further investigate whether EZH2 exerted its functional role in breast cancer CDDP resistance through regulating miR-381 expression, MCF-7/CDDP and MDA-MB-231/CDDP cells were transfected with si-con, si-EZH2, si-EZH2+anti-miR-con or si-EZH2+anti-miR-381. Transfection of si-EZH2 up-regulated miR-381 expression in MCF-7/CDDP and MDA-MB-231/CDDP cells, which was extremely reversed by miR-381 inhibition (Figure 5A and B). MTT assay revealed that down-regulation of EZH2 facilitated CDDP sensitivity of MCF-7/CDDP and MDA-MB-231/CDDP cells, nevertheless, the inductive effect of EZH2 inhibition on CDDP sensitivity of MCF-7/CDDP and MDA-MB-231/CDDP cells was patently abolished by miR-381 down-regulation (Figure 5C and D). Furthermore, introduction of anti-miR-381 extremely demolished the inductive effect of silenced EZH2 on apoptosis in MCF-7/CDDP and MDA-MB-231/CDDP cells (Figure 5E and F). Collectively, these results suggested that EZH2 inhibition contributed to CDDP resistance of breast cancer cells through epigenetically silencing miR-381.
EZH2 Knockdown Improves CDDP Sensitivity In Tumors In Vivo
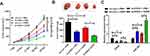
To confirm whether EZH2 knockdown could possibly increase CDDP’s cytotoxic impact, a breast cancer mouse xenograft model was created. We made an sh-EZH2 stably expressing MCF-7/CDDP cell line and inoculated them subcutaneously into nude mice. Thenceforth, mice were injected intraperitoneally with PBS or CDDP, and tumor size was evaluated every 7 days. Xenograft tumors from EZH2-silencing MCF-7/CDDP cells grew evidently slower than tumors from sh-con-transfecting MCF-7/CDDP cells. CDDP treatment suppressed tumors’ growth from sh-con-transfecting MCF-7/CDDP cell. Furthermore, slower tumor growth was displayed in tumors from EZH2-silencing MCF-7/CDDP cells than that from sh-con-transfecting MCF-7/CDDP cells, following CDDP administration (Figure 6A). At the 35th day post-treatment, mice were euthanized and xenograft tumors were excised and weighed. In comparison with the sh-con + PBS group, the sh-con + CDDP group and the sh-EZH2 + PBS group displayed light tumor weight. Moreover, tumors from EZH2-silencing MCF-7/CDDP cells were lighter than that from sh-con-transfecting MCF-7/CDDP cells after CDDP treatment (Figure 6B). Besides, CDDP treatment or EZH2 knockdown inhibited EZH2 expression while up-regulated miR-381 expression, which was strengthened by the combination of EZH2 silencing and CDDP exposure (Figure 6C). All these data suggested that knockdown of EZH2 improved CDDP sensitivity in breast cancer cells in vivo.
Discussion
Therapeutic outcome has been restricted by chemoresistance severely for breast cancer patients. Consequently, to reveal the underlying mechanism and uncover novel therapeutic strategies for chemoresistance is not dispensable. In this work, we found that EZH2 expression was strikingly increased in CDDP-resistant breast cancer tissues and cells. Moreover, EZH2 knockdown re-sensitized CDDP-resistant MCF-7/CDDP and MDA-MB-231/CDDP cells to CDDP. Prominently, EZH2 silencing heightened the response of breast cancer cells towards CDDP through epigenetically suppressing miR-381 expression. Therefore, EZH2 could contribute to breast cancer CDDP resistance and EZH2 inhibition may be an effective scheme for CDDP chemoresistance in breast cancer.
Elucidating the molecular mechanism underlying CDDP resistance was helpful to discover reasonable and effective targeted therapeutic strategies to overwhelmed CDDP resistance. Our study found that EZH2 expression was raised in CDDP-resistant breast cancer tissues and cells, and EZH2-silencing enhanced the responder of CDDP-resistant breast cancer cells towards CDDP. In line with our findings, increasing evidence demonstrated that dysregulated EZH2 was implicated with CDDP resistance in various malignancies. For example, down-regulated EZH2 mediated by miR-124 could inhibit cell proliferation, induce cell apoptosis, and sensitize gastric cancer cells to CDDP.25 EZH2 was overexpressed in hepatocellular carcinoma cells, and EZH2 knockdown improved the sensitivity of Bel/CDDP cells to CDDP by inhibiting multidrug resistance protein 1(MDR1) expression.26,27 Moreover, EZH2 up-regulation led to CDDP resistance through inhibiting DPYD expression.28 Furthermore, down-regulation of EZH2 facilitated the sensitivity of A2780/DDP cells to cisplatin in epithelial ovarian cancer through suppressing BRCA1 expression.29 All these findings indicated that targeted inhibiting EZH2 maybe a hopeful therapeutic strategy for CDDP chemoresistance.
How elevated EZH2 helps to CDDP resistance in breast cancer remains elusive. Hence, the operative mechanism of EZH2 was further explored in this work. Emerging evidence suggested that EZH2 transcriptionally inhibited the expression of miRNAs through increasing H3K27me3 on their promoter region.22,23 Moreover, miR-381 has been reported to be down-regulated in cancers attributed to DNA hypermethylation.15 However, whether EZH2 could regulate miR-381 expression at transcriptional level in breast cancer continued to be indescribable. In our study, Chipbase database prediction and ChIP and luciferase reporter assays verified that EZH2 could bind with miR-381 and transcriptionally suppress miR-381 expression in MCF-7/CDDP and MDA-MB-231/CDDP cells. Mounting evidence has indicated that miR-381 played a tumor suppressor role in tumorigenesis.17,30–32 Moreover, down-regulated miR-381 was related to chemoresistance in many cancers.33–36 Especially, miR-381 overexpression could suppress proliferation, and metastasis in breast cancer cells through targetly suppressing CXCR4.37 Prominently, miR-381 could improve the response of non-small cell lung cancer cells to CDDP through the inactivation of nuclear factor-ĸB signaling.38 A recent study reported that miR-381 could overcome CDDP resistance in breast cancer through directly inhibiting MDR1.20 Correspondingly, our found indicated that miR-381 overexpression overcame CDDP resistance in MCF-7/CDDP and MDA-MB-231/CDDP cells. Additionally, miR-381 inhibition reversed EZH2 knockdown-mediated enhancement of CDDP sensitivity in MCF-7/CDDP and MDA-MB-231/CDDP. All these data demonstrated that EZH2 inhibition re-sensitized CDDP-resistant breast cancer cells towards CDDP through epigenetically silencing miR-381 in breast cancer.
In conclusion, our study demonstrated that EZH2 knockdown improved CDDP sensitivity of breast cancer cells through epigenetically silencing miR-381, providing a promising therapeutic target for breast cancer CDDP resistance.
Acknowledgment
This work was supported by Henan Science and Technology program.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca A Cancer J Clin. 2016;66:1. doi:10.3322/caac.21332
2. Sutter SA, Slinker A, Balumuka DD, Mitchell KB. Surgical management of breast cancer in Africa: a continent-wide review of intervention practices, barriers to care, and adjuvant therapy. J Glob Oncol. 2017;3(2):162–168. doi:10.1200/JGO.2016.003095
3. Villarreal-Garza C, Khalaf D, Bouganim N, et al. Platinum-based chemotherapy in triple-negative advanced breast cancer. Dtsch Med Wochenschr. 2015;26(8):894–901.
4. Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi:10.1038/nrd1984
5. Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34(4):378. doi:10.1016/j.ctrv.2008.01.007
6. Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;300(5616):131.
7. Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi:10.1016/j.gde.2004.02.001
8. Yang CC, Labaff A, Wei Y, et al. Phosphorylation of EZH2 at T416 by CDK2 contributes to the malignancy of triple negative breast cancers. Am J Transl Res. 2015;7(6):1009–1020.
9. Tamgue O, Chai CS, Hao L, et al. Triptolide inhibits histone methyltransferase EZH2 and modulates the expression of its target genes in prostate cancer cells. Asian Pac J Cancer Prev Apjcp. 2013;14(10):5663–5669. doi:10.7314/apjcp.2013.14.10.5663
10. Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;24(6):566–567.
11. Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi:10.1073/pnas.1933744100
12. Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8(1):59. doi:10.7150/ijbs.8.59
13. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nature Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
14. Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi:10.1016/j.tibs.2010.03.009
15. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
16. Liu X, Luo X, Wu Y, et al. MicroRNA-34a attenuates paclitaxel resistance in prostate cancer cells via direct suppression of JAG1/Notch1 axis. Cell Physiol Biochem. 2018;50(1):261–276. doi:10.1159/000494004
17. Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–433. doi:10.1016/j.biopha.2016.12.051
18. Huang R-S, Zheng Y-L, Zhao J, Chun X. microRNA-381 suppresses the growth and increases cisplatin sensitivity in non-small cell lung cancer cells through inhibition of nuclear factor-κB signaling. Biomed Pharmacother. 2018;98:538–544. doi:10.1016/j.biopha.2017.12.092
19. Mi HL, Wang XC, Wang F, et al. miR-381 induces sensitivity of breast cancer cells to doxorubicin by inactivation of MAPK signaling via FYN. Eur J Pharmacol. 2018;839:66–75. doi:10.1016/j.ejphar.2018.09.024
20. Yi DD, Xu L, Wang R, et al. miR-381 overcomes cisplatin resistance in breast cancer by targeting MDR1. Cell Biol Int. 2019;43:12–21. doi:10.1002/cbin.11071
21. Wang WJ, Yao Y, Jiang LL, et al. Knockdown of lymphoid enhancer factor 1 inhibits colon cancer progression in vitro and in vivo. PLoS One. 2013;8(10):e76596. doi:10.1371/journal.pone.0076596
22. Cisneros-Soberanis F, Andonegui MA, Herrera LA. miR-125b-1 is repressed by histone modifications in breast cancer cell lines. Springerplus. 2016;5(1):959. doi:10.1186/s40064-016-2475-z
23. Chen S, Pu J, Bai J, et al. EZH2 promotes hepatocellular carcinoma progression through modulating miR-22/galectin-9 axis. J Exp Clin Cancer Res. 2018;37(1):3. doi:10.1186/s13046-017-0670-6
24. Li J, Ying Y, Xie H, et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. Faseb J. 2018;33(1):1374–1388. doi:10.1096/fj.201800667R
25. Xie L, Zhang Z, Tan Z, et al. MicroRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol Cell Biochem. 2014;392(1–2):153–159. doi:10.1007/s11010-014-2028-0
26. Tang B, Zhang Y, Liang R, Gao Z, Sun D, Wang L. RNAi-mediated EZH2 depletion decreases MDR1 expression and sensitizes multidrug-resistant hepatocellular carcinoma cells to chemotherapy. Oncol Rep. 2013;29(3):1037–1042. doi:10.3892/or.2013.2222
27. Zhang Y, Liu G, Lin C, Liao G, Tang B. Silencing the EZH2 gene by RNA interference reverses the drug resistance of human hepatic multidrug-resistant cancer cells to 5-Fu. Life Sci. 2013;92(17–19):896–902. doi:10.1016/j.lfs.2013.03.010
28. Wu R, Nie Q, Tapper EE, et al. Histone H3K27 trimethylation modulates 5-fluorouracil resistance by inhibiting PU.1 binding to the DPYD promoter. Cancer Res. 2016;76:21. doi:10.1158/0008-5472.CAN-16-0584
29. Li T, Cai J, Ding H, Xu L, Yang Q, Wang Z. EZH2 participates in malignant biological behavior of epithelial ovarian cancer through regulating the expression of BRCA1. Cancer Biol Ther. 2014;15(3):271–278. doi:10.4161/cbt.27306
30. Tian C, Li J, Ren L, Peng R, Chen B, Lin Y. MicroRNA-381 serves as a prognostic factor and inhibits migration and invasion in non-small cell lung cancer by targeting LRH-1. Oncol Rep. 2017;38(5):3071–3077. doi:10.3892/or.2017.5956
31. Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi:10.1186/s13046-017-0499-z
32. He X, Wei Y, Wang Y, Liu L, Wang W, Li N. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231.
33. Wang Z, Yang J, Xu G, et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6(5):3147–3164. doi:10.18632/oncotarget.3061
34. Chen B, Duan L, Yin G, Jing T, Jiang X. miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J Chemother. 2013;25(4):229–238. doi:10.1179/1973947813Y.0000000092
35. Li Y, Zhao C, Yu Z, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7(42):68585–68596. doi:10.18632/oncotarget.11861
36. Xu Y, Ohms S, Li Z, et al. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8(11):e82062. doi:10.1371/journal.pone.0082062
37. Xue YB, Xu WJ, Zhao W, et al. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–433. doi:10.1016/j.biopha.2016.12.051
38. Huang RS, Zheng YL, Zhao J, Chun X. microRNA-381 suppresses the growth and increases cisplatin sensitivity in non-small cell lung cancer cells through inhibition of nuclear factor-κB signaling. Biomed Pharmacother. 2017;98:538. doi:10.1016/j.biopha.2017.12.092
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.