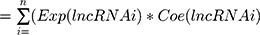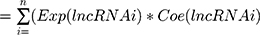Back to Journals » OncoTargets and Therapy » Volume 13
Extracellular Matrix–Related Six-lncRNA Signature as a Novel Prognostic Biomarker for Bladder Cancer
Authors Qing L, Gu P, Liu M, Shen J, Liu X, Guang R, Ke K, Huang Z, Lee W , Zhao H
Received 29 September 2020
Accepted for publication 10 November 2020
Published 7 December 2020 Volume 2020:13 Pages 12521—12538
DOI https://doi.org/10.2147/OTT.S284167
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Liangliang Qing,1,* Peng Gu,1,* Mingsheng Liu,2 Jihong Shen,1 Xiaodong Liu,1 Runyun Guang,1 Kunbin Ke,1 Zhuo Huang,1 Wenhui Lee,1,3 Hui Zhao1
1Department of Urology, First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, People’s Republic of China; 2Second Ward of Urology, Qujing Affiliated Hospital of Kunming Medical University, Qujing, Yunnan, People’s Republic of China; 3Key Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences, Kunming Institute of Zoology, Kunming, Yunnan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenhui Lee
Key Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences, Kunming Institute of Zoology, 32 Jiaochang East Road, Kunming 650223, Yunnan, People’s Republic of China
Email [email protected]
Hui Zhao
Department of Urology, First Affiliated Hospital of Kunming Medical University, 295 Xichang Road, Kunming 650032, Yunnan, People’s Republic of China
Email [email protected]
Introduction: Bladder cancer (BC) is the fourth-commones cancer and the sixth-leading cause of cancer-related death among men. However, a lack of reliable biomarkers remains a problem forprognosis and treatment of BC. lncRNAs have been shown to play important roles in various cancers, and have emerged as promising biomarkers for cancer prognosis and treatment.
Methods: In this study, using univariate and multivariate Cox regression analysis, we examined the differential expression profiles of 1,651 lncRNAs in the TCGA BLCA cohort and created a prognostic gene signature composed of six lncRNAs (for SNHG12, MAFG-DT, ASMTL-AS1, LINC02321, LINC01322, and LINC00922), designed the SMALLL signature.
Results: The SMALLL signature displayed significant prognostic power for overall survival for BC patients in multiple cohorts. Gene Ontology analysis showed that genes coexpressed with the SMALLL signature were associated with the extracellular matrix network, and immune cell–infiltration analysis showed that activated naïve B cells, regulatory T cells, M0 macrophages, eosinophils, resting memory CD4 T cells and resting NK cells were significantly different in high- and low-risk groups. We also confirmed differential expression of the lncRNAs of the SMALLL signature in BC tissue and paracancer normal tissue by qRT-PCR analysis. Cell-invasion and -migration experiments showed that MAFG-AS1, ASMTL-AS1, LINC02321, and LINC00922 significantly affected cell invasion and migration.
Conclusion: Our study revealed that the lncRNA signature is an important predictive factor of prognosis and provides a promising biomarker for BC.
Keywords: long noncoding RNA, bladder cancer, extracellular matrix, immune-cell infiltration, prognosis, signature
Introduction
Bladder cancer (BC) is the fourth-commonest cancer and the sixth-leading cause of cancer-related death among men.1 With regard to histological characteristics, nearly 90% of BCs are urothelial carcinoma, of which approximately 75%–85% are classified as non–muscle invasive BC (NMIBC) and the rest MIBC.2 At present, treatment options for BC include surgery,3 radiotherapy, chemotherapy,4 immunotherapy,5 and recently developed immunocheckpoint inhibitors.6 In general, the American Joint Committee on Cancer TNM-staging system is used to guide prognosis and management options for BC patients. However, although patients with NMIBC have favorable survival after initial treatment, 5-year recurrence and 5-year progression remain as high as 70% and 30%, respectively.7,8 As such, there is an urgent need for development of effective prognostic biomarkers for earlier detection and precise recurrence prediction of BC.
lncRNAs are RNA transcripts that are longer than 200 bases and lack translational potential.9 Although the functions of lncRNAs have not been fully elucidated, studies have shown that lncRNAs play an important role in dose-compensation effects, epigenetic regulation, cell proliferation and differentiation, chromatin modification, immunity, and other biological processes.10,11 Mounting evidence indicates that the deregulation of lncRNA is implicated in the tumorigenesis and progression of various types of cancers, including BC.12,13 For example, UCA1 lncRNA is upregulated in BC.14 UCA1 overexpression promotes cell-cycle progression, carcinogenesis, and cancer invasion of BC cells through enhancing ERK1/2, MAPK, and PI3K–Akt pathways.15 H19 is another well characterized lncRNA that is highly expressed in BC and many other cancers.16,17
Lack of an effective prediction approach is one of the main causes of poor prognosis of BC patients. Emerging evidence has suggested that lncRNAs may serve as novel biomarkers for accurate prognosis of cancer patients.18 Various studies have shown that lncRNA expression is significantly associated with survival in patients with ovarian cancer,19 papillary thyroid carcinoma,20 pancreatic cancer,21 non–small cell lung cancer,22 and esophageal cancer.23 Recently, a set of immunorelated lncRNAs was found to be significantly correlated with progression of diverse cancer types, including hepatocellular carcinoma,24 breast cancer,25 non–small cell lung cancer,26 anaplastic gliomas,27 glioblastoma multiforme,28 and diffuse large B–cell lymphoma.29 In addition, multiple lncRNA-based gene signatures have been developed to predict recurrence in hepatocellular carcinoma,30 breast cancer,31 and colorectal cancer,32 as well as BC.33–35 These findings indicate better efficacy of lncRNA signatures in predicting prognosis than individual lncRNAs.
Changes in the composition, physical properties, and spatial conformation of the extracellular matrix (ECM) play an important role in the process of tumor proliferation and invasion. Witkowski et al found that there is extensive immunomicroenvironment remodeling in B-cell acute lymphoblastic leukemia.36 Varn et al found that there are complex interactions between different immune-cell types in the tumor microenvironment and proved that these interactions have a significant impact on patient survival.37 Through the pathway enrichment analysis of genes related to SMALL (SNHG12, MAFG-DT, ASMTL-AS1, LINC02321, LINC01322, and LINC00922) characteristics, we found that these genes significantly enriched pathways related to the ECM, including ECM structural constituents, cell adhesion–molecule binding, actin binding, and glycosaminoglycan binding.
In this study, we developed a novel lncRNA signature for survival prediction of BC patients, with increased patient samples and improved methods. We conducted a comprehensive analysis of lncRNA-expression profiles in a cohort of 372 BC patients from the Cancer Genome Atlas (TCGA) database, and identified and validated a six-lncRNA signature to predict overall survival (OS) of patients with BC. In addition, we verified the accuracy of the signature for survival prognosis in two external data sets (GSE31684 and GSE32894), and compared the signature we established with other existing BC lncRNA prognostic signatures.
Methods
BC Data-Set Retrieval and Differential Gene-Expression Analysis
To screen lncRNAs that can be used as effective prognostic markers for BC patients, gene-expression profiles and clinical data of the TCGA Urothelial Bladder Carcinoma (BLCA) cohort were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov). The TCGA BLCA cohort comprises 429 cases, and detailed information is available on the portal. Gene-expression profiles and survival data of GSE31684 and GSE32894 were downloaded from the Gene Expression Omnibus(https://www.ncbi.nlm.nih.gov/geo).
After transformation of Ensembl IDs to gene symbols, mRNA and lncRNA profiles were extracted for differential analysis using the R program. Samples were sorted into a tumor group and a normal group. Then the EdgeR program was used to normalize the data and determine differentially expressed genes. Briefly, the estimateCommonDisp and estimateTagwiseDisp functions were used to calculate ordinary dispersion and dispersion within the gene range, respectively, then the exactTest function was used to perform Fisher’s exact test. Finally, the topTags function was used to extract differentially expressed genes. In order to analyze correlations between expression characteristics of lncRNA and OS and recurrence of BC patients, 372 BC cases with complete clinical information were included in this study. These were randomly divided into a training group and a validation group. Detailed clinical features of all groups were listed in Table 1.
 |
Table 1 Clinical Features of BC Patients in Training and Validation Groups |
Risk-Score Calculation
Univariate Cox proportional-hazard regression analysis was carried out for the differentially expressed lncRNAs to screen the candidates that significantly correlated with OS of BC patients. Next, correlation coefficients of these candidate lncRNAs were generated by multivariate Cox regression analysis. Expression of the selected lncRNAs was weighted linearly according to their coefficients, and a risk-score formula was obtained:
where i is the identifier of selected lncRNAs, n the number of lncRNA genes in prognosis, Exp the expression value of lncRNA, and Coe the estimated regression coefficient of lncRNA on multivariate Cox proportional-hazard regression analysis. This risk-scoring model fully considers the function of each prognostic lncRNA gene. Each patient received a risk score: a linear combination of significant levels of lncRNA expression weighted by their respective Cox regression coefficients.
Pathway-Enrichment Analysis and Possible Mechanism Exploration
All coexpressed genes were selected for pathway enrichment analysis. The R package ClusterProfiler38 was used for enrichment. The threshold of significance of false positive ratefor biological processes and pathway of significant enrichment was set at 0.05. GSEA version 4.0.239,40 was used for biological processes and pathway-enrichment analysis. The threshold of significance was set as 0.05, and normalized enrichment score (NES) was set at |NES|>1. We introduced coexpressed genes with correlation coefficients >0.6 into STRING (http://string-db.org) to establish a PPI network. Next, we imported the PPI network into Cytoscape and used the CytoHubba plug-in to find the hub genes.
Immune Cell–Infiltration Analysis
As an important part of the tumor microenvironment, immune cells play an important role in tumor growth and metastasis.36 Therefore, we performed an in silico deconvolution of 22 immune-cell types through the CIBERSORT algorithm (https://cibersort.stanford.edu).41 After deleting samples with no statistical significance, 304 samples remained, and these were split into high-risk (n=153) and low-risk (n=151) cohorts by the risk model. A nonparametric Wilcox test was performed to assess whether the infiltration of immune cells differed between the high-risk and low-risk groups. A violin plot was selected to show results, in which blue represented the low-risk cohort and red the high-risk cohort.
lncRNA-Expression Analysis in Human BC Tissue
All the BC tissue samples (n=20) were obtained by surgery with the written consent of patients who underwent surgery at the First Affiliated Hospital of Kunming Medical University. All samples were pathologically confirmed as urothelial carcinoma by two pathologists independently. Ethics consent was approved by the First Affiliated Hospital of Kunming Medical University’s Committees for Ethical Review of Research Involving Human Subjects. Total RNA was extracted with the Eastep Super Total RNA-extraction kit, followed by cDNA synthesis using the GoScript reverse-transcription system (Promega). Expression of lncRNAs was determined by qPCR using Go-taq qPCR Master Mix (Promega). Primers (Table 2) designed using Primer3 (http://primer3.ut.ee), and DNA oligos were synthesized by Xinfan Biotechnology, Shanghai, China.
 |
Table 2 Nucleotide sequence of primers for lncRNA qPCR |
shRNA Interference
DMEM, FBS, penicillin–streptomycin, trypsin–EDTA and PBS were purchased from Biological Industries (Kibbutz Beit Haemek, Israel). SVHUC1, RT4, T24 and UMUC3 cell lines were purchased from Kunming Institute of Zoology (Chinese Academy of Sciences, Kunming, China). Cell cultures were routinely grown at 37°C with 5% CO2 using DMEM + 10% FBS + 1% penicillin–streptomycin solution. Plasmids of shRNA targeting MAFG-AS1 and negative shRNA were designed and synthesized by Shanghai Jikai Gene Technology (GenePharma). The shRNA sequence was CAGGGCAATTCCAACCAAGAA. Transfections were performed according to the protocol provided by the manufacturer (Lipo3000, Thermo Fisher Scientific). Cell-invasion and -migration assays were performed using the Cultrex (Trevigen, MD, USA) BME.
Statistical Analysis
To discover potential factors affecting the prognosis of BC patients, univariate Cox proportional-hazard regression was applied to determine associations between lncRNA expression and OS in the training group. If corrected P<0.05, corresponding lncRNAs were statistically significant and considered as candidate prognostic lncRNAs of BC. Multivariate Cox proportional-hazard regression was carried out among the pool of candidate prognostic lncRNAs, and those with corresponding P<0.005 were identified as optimal prognostic lncRNAs impacting the survival of BC patients.
An individual risk score for each patient was built for predicting prognosis by including expression of each optimal prognostic lncRNA, weighted by their estimated regression coefficients on multivariate Cox regression model:
The risk-score model was a measure of prognostic risk for each BC patient. Based on the median risk score of the training group as the cutoff, patients with BC were sorted into two groups: high risk and low risk. A high risk score indicated poor survival for BC patients.
In each group, we validated the reliability and validity of the risk-score formula. Kaplan–Meier analysis was utilized to compare the survival rate of the groups. The survival difference between the low-risk and high-risk groups was assessed by the log-rank test. Time-dependent ROC-curve analysis for OS was used to display the performance of the lncRNA risk model. Univariate and multivariate analyses with Cox proportional-hazard regression for OS were performed on individual clinical risk factors with and without the six-lncRNA signature in each cohort. HRs and 95% CIs were estimated.
In addition, multivariate Cox proportional-hazard analysis and stratified data analysis were used to test whether the risk score was independent of other clinical features, with age, sex, race, pathological grade, TNM stage, and recurrence status as covariates. P<0.05 was considered statistically significant.
All the analyses were implemented in SPSS version 24.0 or R version 3.6.1 with the following packages:“Limma”, “EdgeR”, “Pheatmap”, “Caret”, “Survival”, “Magrittr”, “Survminer”, “SurvivalROC”, “RMS”, “PreprocessCore”, “Cibersort”, “Vioplot”, “org.Hs.eg.db”, “ClusterProfiler”, “ggpubr”, and “ggplot2”. All the hypotheses were two-sided, and P < <0.05 was considered statistically significant.
Results
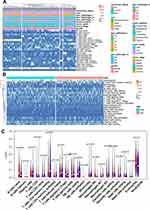
The TCGA BLCA cohort (n=372) was randomly divided into a training group (n=188) and a validation group (n=184). There were no significant differences in survival time, survival state, age, sex, race, pathological grade, disease stage, T stage, M stage, or recurrence between the two groups, but for N stage, P<0.05 (Table 1). To determine the correlation between of lncRNA expression and the OS rate of BC patients, we carried out a variation analysis of mRNA and lncRNA expression, respectively, in the training group (Figure 1A, B). Next, we analyzed the differentially expressed lncRNAs in the training group by univariate Cox regression analysis. Interestingly, we found thatexpression levels of several lncRNAs were closely related to OS of the patients (P<0.05). Then, multivariate Cox proportional-hazard regression analysis resulted in six top candidates for a gene signature, designated the SMALLL signature, based on the first letter of the component genes: SNHG12, MAFG-AS1, ASMTL-AS1, LINC02321, LINC01322, and LINC00922. Among the six lncRNAs, positive coefficients indicated that expression of MAFG-AS1, LINC02321, LINC01322, and LINC00922 correlated with longer OS, whereas expression of SNHG12 and ASMTL-AS1 inversely correlated with survival of patients (Table 3).
 |
Table 3 Six lncRNAs Significantly Associated With OS of BC Patients in the Training Group |
To assess whether the combination of the identified lncRNAs improved power in predicting OS of BC patients, we established a risk-scoring formula for OS prediction according to expression of the six lncRNAs: risk score=(−0.301151077 × expression value SNHG12) + (0.316595657 × expression value MAFG-AS1) + (0.172517499 × expression value LINC02321) + (0.179091241 × expression value ASMTL-AS1) + (0.158178271 × expression value LINC01322) + (0.124204122 × expression value LINC00922). Then the risk scores of the SMALLL signature were calculated for each patient in the training group. Next, we ranked the patients according to their risk scores and divided them into high-risk (n=94) and low-risk (n=94) groups, with the median risk score of the training group as the cutoff value (Figure 1C). As expected, OS of BC patients with higher risk scores (39.36%) was shorter than that of BC patients with lower risk scores (77.02%, Figure 1E). In addition, expression of high-risk lncRNAs (MAFG-AS1, LINC02321, LINC01322, and LINC00922) was higher in the high-risk group, while there was no significant difference in expression of low-risk lncRNAs (SNHG12 and ASMTL-AS1) between the high-risk and low-risk groups (Figure 1G). To validate the prognostic performance of the SMALLL signature, we evaluated its prognostic power in the validation group derived from the TCGA BLCA cohort. Using the risk-score formula, patients in the validation group were divided into high-risk (n=92) and low-risk (n=92, Figure 1D) groups. OS of BC patients and expression patterns of the six lncRNAs were similar to the training group (Figure 1F and H).
SMALLL Signature Significantly Predicted OS of BC Patients
Kaplan–Meier analysis showed that OS of the high-risk group was significantly lower than the low-risk group (HR 2.89, 95% CI 0.24–0.58; P=4.626–66, Figure 2A) in the training group (n=188). The OS rate of the high-risk group was 59.1% in 24 months, 38.9% in 48 months, 31.9% in 72 months, 25.8% in 96 months, and 85.6%, 69.1%, 60.6% and 57.9% in the low-risk group, respectively. A similar effect was also observed in the validation group (Figure 2B). To further test the utility of this signature, we examined its prognostic performance in the survival of BC patients in two additional publicly available BC cohorts: the GSE3168442 and GSE3289443 data sets. The results showed that the survival rate in the high-risk group was significantly lower than the low-risk group, with P=3.677–4 (GSE31684, Figure 2C) and 3.552–3 (GSE32894, Figure 2D), respectively. Three distinct lncRNA gene signatures have been reported to have significant prognostic performance on survival of BC patients.34,44,45 The composition of our signature is different from these published signatures. In addition, ROC analysis showed that the SMALLL signature performed better than the three lncRNA signatures (Figure 2E).
Survival Prediction by SMALLL Signature Was Independent of Classic Clinical Factors
We conducted multivariate Cox proportional-hazard regression analysis and multi-ROC analysis to assess whether the SMALLL signature predicted OS of BC independently of classic clinical and pathological factors. Selected covariates included patient age, sex, race, pathological grade, TNM stage, and recurrence status (Tables 4 and 5). Our results showed that the prognostic risk score for the SMALLL signature (HR 1.797, 95% CI 1.350–2.393; P=0.001) was independent of these clinical features in the training group (Figure 3A). Consistently, similar results were obtained in the validation group and all samples: validation group — high-risk group vs low-risk group (HR 1.002, 95% CI 0.717–1.401; P=0.05); all samples — high-risk group vs low-risk group (HR 1.662, 95% CI 1.293–2.137; P<0.001, Figure 3B and C). We noticed that in the validation group, the statistical significance of the multivariate analysis was marginal (Figure 3B, right panel). The variation might have been due to limited sample size, as the statistical significance improved markedly when all samples were included (Figure 3C, right panel). To compare sensitivity and specificity between various clinical parameters and the SMALLL signature in OS prediction for BC patients, we carried out ROC analysis. As shown in Figure 3D, the predictive capability of the SMALLL signature (AUC 0.753) had greater predictive power than the classic clinical parameters (AUC 0.514–0.671).
 |
Table 4 Univariate and Multivariate Cox Regression Analysis on Associations between SMALLL Signature and OS of BC Patients in Training Group |
 |
Table 5 Univariate and Multivariate Cox Regression Analysis on Associations between SMALLL Signature and OS of BC Patients in Validation Group |
Functional Enrichment Analysis and Possible Mechanism Exploration
To determine potential biological roles involved in the SMALLL signature, we performed a functional enrichment analysis for coexpression of genes. Gene Ontology (GO) analysis showed that these genes were significantly enriched in cancer-related networks, including ECM structural constituents, cell adhesion–molecule binding, focal adhesion, and regulation of actin cytoskeleton (Figure 4A). Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that these genes were significantly enriched in focal adhesion, regulation of actin cytoskeleton, the Fanconi anemia pathway, ECM-receptor interaction, and DNA replication (Figure 4B). GSEA results showed that the transcriptional profile of the high-risk group was positively correlated with the pathways of adherens-junction organization (P<0.05, |NES|>1), cell-matrix adhesion (P<0.05, |NES|>1), cell substrate–junction assembly (P<0.05, |NES|>1), glycoprotein-metabosm processes (P<0.05, |NES|>1), and response to inorganic substances (P<0.05, |NES|>1; Figure 4C–G). Then, we found hub genes through the PPI network of co-expressed genes. The top-nine hub genes — COL3A1, COL1A2, COL1A1, COL5A1, COL6A1, COL5A2, COL6A2, COL6A3, and COL12A1 — all belong to the gene family encoding collagen (Figure 4H). These results indicated that the lncRNAs of the SMALLL signature might be functionally implicated in the regulation of ECM networks.
Immune Cell–Infiltration Analysis
By analyzing the abundance of immune-cell infiltration for different clinical characteristics and different risk groups, we found that the abundance had a greater correlation with tumor grade, T staging, and M staging and less correlation with age, sex, recurrence status, tumor location, and N staging (Figure 5A). Immune cell–infiltration abundance in the low-risk group was significantly higher than the high-risk group (Figure 5B). After analyzing each immune cell individually, it was found that naïve B cells, regulatory T cells, M0 macrophages, and eosinophils infiltrated more abundantly in the high-risk group, while resting memory CD4 T cells and resting NK cells infiltrated more abundantly in the low-risk group (Figure 5C).
Expression of lncRNAs in SMALLL Signature
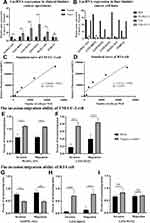
To further validate our findings, we measured the expression of the six lncRNAs in cancer and paracancerous tissue in 20 patients with BC by qPCR. Our data showed that expression of high-risk lncRNAs (MAFG-AS1, LINC02321, LINC01322, and LINC00922) was significantly higher in BC tissue than paracancerous bladder tissue. Consistently, expression of low-risk lncRNAs (SNHG12 and ASMTL-AS1) was higher in paracancerous BC tissue (Figure 6A).
Next, we measured expression of the six lncRNAs in four BC cell lines (RT4, T24, UMUC3, and SVHUC1) by qPCR. The results showed that ASMTL-AS1, LINC00922, and LINC01322 were highly expressed in RT4 cells and LINC02321, MAFG-AS1, and SNHG12 highly expressed in UMUC3 cells (Figure 6B). Therefore, RT4 and UMUC3 cells were used for RNA-interference experiments.
Standard growth curves were examined for UMUC3 and RT4 cells (Figure 6C and D). As GO and KEGG analyses indicated connection of lncRNAs in the SMALLL signature in the ECM network, often implicated in cancer metastasis, and the difference in SNHG12 between cancer and adjacent cancer was not significant. Therefore, we assessed the effect of MAFG-AS1, ASMTL-AS1, LINC01322, LINC02321, and LINC00922 depletion on the invasion and migration of BC cells. To this end, knockdown of MAFG-AS1 and LINC02321 in UMUC3 cells was achieved by transient transfection of shRNA-expressing plasmids. Knockdown of ASMTL-AS1, LINC00922, and LINC01322 in RT4 cells was achieved in the same way. Two days after transfection, cell migration and invasion were determined by transwell based in vitro assays, using calcein AM for cell quantification. As shown in Figure 6E–I, knockdown of MAFG-AS1, LINC02321, and LINC00922 significantly inhibited the migration and invasion of UMUC3 and RT4 cells, and knockdown of ASMTL-AS1 significantly promoted the migration and invasion of RT4 cells. The knockdown of LINC01322 had no significant effect on the migration and invasion of RT4 cells. These results suggested that MAFG-AS1, LINC02321, and LINC00922 overexpression may promote invasiveness of BC cells and low expression of LINC01322 has a similar effect. However, the mechanism of how those lncRNAs contribute to BC-cell invasion remains unknown, and further investigations are needed to elucidate it.
Discussion
Traditionally, treatment and prognosis of BC patients are based on pathological grading and imaging results. However, the risk of recurrence and cancer-related death remains high, even after radical cystectomy, postoperative radiotherapy, and chemotherapy,46,47 especially in MIBC patients. As such, traditional TNM staging and imaging assessment are unable to predict the invasiveness of BC and determine prognoses for BC patients properly. Therefore, it is very important to find new and effective prognostic biomarkers for BC patients.
Accumulating evidence has indicated that lncRNAs are involved in many physiological processes, and deregulated lncRNAs may contribute to the occurrence, development, and metastasis of cancer.48,49 In addition, lncRNAs have shown great potential as novel molecular biomarkers in early diagnosis, treatment, and prognosis in many types of cancer.50–52 In recent years, many tissue-specific lncRNAs have been found in normal tissue by transcriptional profiling, and lncRNA dysregulation has been found in a variety of human cancers.13,53,54 Studies have shown that expression of some lncRNAs can predict the risk of cancer progression in patients with different types of cancer, such as breast cancer,31 colorectal cancer,32 and ovarian cancer.19 However, due to the complexity of the lncRNA-transcription landscape, a single lncRNA may not accurately predict the prognosis of cancer patients. Although attempts to use lncRNAs in prognoses for BC patients have been described,55,56 further exploration and improvement is required in this field. By comprehensive mining of lncRNA-expression profiles in the TCGA cohort, we established a novel six-lncRNA signature, the SMALLL signature, that was significantly related to the prognosis of BC patients.
To the best of our knowledge, little is known about the implications of the six lncRNAs in the SMALLL signature in human cancers, except for SNHG12. SNHG12 is a small nucleolar host gene that has been described in several types of cancer including breast,57 gastric,58 esophagus,59 liver,60 and prostate cancer.61 Upregulation of SNHG12 has shown proliferative and antiapoptotic effects in some cancer-cell types, and altered expression of SNHG12 correlates with prognosis and survival of cancer patients.62 Additionally, SNHG12 is also differentially expressed in normal and malignant bladder tissue.63 For the other five lncRNAs, ASMTL-AS1 and LINC00922 have only recently been described in papillary thyroid cancer64 and lung cancer.65 ASMTL-AS1 downregulation in papillary thyroid cancer is positively linked to advanced clinical stage and unfavorable outcomes. It has been proposed that ASMTL-AS1 is a tumor suppressor via the miR93-3p–miR660–FOXO1 axis.64 LINC00922 upregulation is associated with poor prognosis in lung cancer and favors cancer-cell proliferation and invasion.65 However, their biological role in BC remains elusive.
Through pathway-enrichment analysis of the lncRNAs in the SMALL signature, we found that these lncRNAs may be involved in regulation of the ECM. Through PPI-network analysis of coexpressed genes, we found some hub genes, of which the first nine hub (COL3A1, COL1A2, COL1A1, COL5A1, COL6A1, COL5A2, COL6A2, COL6A3, and COL12A1) are members of the gene family involved in collagen coding. This further verified the hypothesis that these lncRNAs may be involved in regulation of the ECM.66 Next, we confirmed through in vitro experiments that knockdown of these lncRNAs significantly affects the invasion and migration of BC cells.
Development and morbidity of BC are associated with various risk factors, such as aging, which correlates with accumulation of gene mutations.67,68 Aging also causes alterations in sex hormones and their receptors, which are associated with the impact of sex differences on the morbidity and development of BC.69 As a result, we propose that this might be the reason that the prognostic value of the SMALLL signature was more efficient in women.
The present study has some innate limitations inevitably. First of all, this study was retrospective because it was based on the TCGA data set and not validated in prospective clinical trials. Secondly, the mechanism behind lncRNAs in our classifiers is still completely unclear. More importantly, the TCGA cohort did not contain integrated information on some important features, such as pharmacological and surgical interventions. Therefore, further research on specific lncRNAs is required. Despite these shortcomings, the results show that our lncRNA-based classifier can be used as a reliable predictor of BC survival and recurrence. In summary, we identified a novel lncRNA signature that is significantly associated with OS in BC patients. Further analysis of the signature indicates that the lncRNAs were enriched in the extracted matrix-structue pathway. Our findings provide insight into an lncRNA signature that might be a prognostic biomarker for BC.
Ethics and Consent
All patients in the study signed informed-consent forms before surgery at the First Affiliated Hospital of Kunming Medical University. Our research was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Kunming Medical University and complied with the ethical principles of the 1964 Declaration of Helsinki and later versions.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants 81802548 and 81860451), Natural Science Foundation of Yunnan Province (grant 202001AW070001), National Natural Science Foundation of Yunnan–Kunming Medical University Joint Foundation (grants 2019FE001[−005], 2019FE001[−064], 2019FE001[−136], and 2014FB031), Project for Innovation Team of Yunnan Provincial Science and Technology Department, China (grant 2018HC005), Scientific Research Project of Yunnan Provincial Educational Department (grant 2018JS208), Funding for Young Doctors (to Peng Gu) from the first Affiliated Hospital of Kunming Medical University (grant 2017BS016), and Yunnan Health Training Project of High Level Talents (to Peng Gu, grant H2018070).
Author Contributions
All authors made a significant contribution to the work reported, whether in conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas, took part in drafting, revising, or critically reviewing the article, gave final approval to the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work, and declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Giridhar KV, Kohli M. Management of muscle-invasive urothelial cancer and the emerging role of immunotherapy in advanced urothelial cancer. Mayo Clin Proc. 2017;92(10):1564–1582. doi:10.1016/j.mayocp.2017.07.010
3. Jewett HJ. Conservative treatment vs radical surgery for superficial cancer of the bladder. JAMA. 1968;206(12):2720–2721. doi:10.1001/jama.1968.03150120054013
4. Mitin T, Hunt D, Shipley WU, et al. Transurethral surgery and twice-daily radiation plus paclitaxel-cisplatin or fluorouracil-cisplatin with selective bladder preservation and adjuvant chemotherapy for patients with muscle invasive bladder cancer (RTOG 0233): a randomised multicentre Phase 2 trial. Lancet Oncol. 2013;14(9):863–872.
5. Cho MJ, Kim MJ, Kim K, et al. The immunotherapeutic effects of recombinant bacillus calmette-guerin resistant to antimicrobial peptides on bladder cancer cells. Biochem Biophys Res Commun. 2019;509(1):167–174. doi:10.1016/j.bbrc.2018.12.097
6. Massari F, Di Nunno V, Cubelli M, et al. Immune checkpoint inhibitors for metastatic bladder cancer. Cancer Treat Rev. 2018;64:11–20.
7. Waseda Y, Kobayashi S, Kanda E, et al. Impact of bladder neck involvement on recurrence in patients with non-muscle-invasive bladder cancer: an analysis based on a time-dependent model. Clin Genitourin Cancer. 2020;18(2):e62–e70
8. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. doi:10.1016/S0140-6736(16)30512-8
9. Mirhosseini SA, Sarfi M, Samavarchi Tehrani S, Mirazakhani M, Maniati M, Amani J. Modulation of cancer cell signaling by long noncoding RNAs. J Cell Biochem. 2019;120(8):12224–12246. doi:10.1002/jcb.28847
10. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
11. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi:10.1016/j.cell.2013.06.020
12. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
13. Peng W-X, Koirala P, Mo -Y-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
14. Wang XS, Zhang Z, Wang HC, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12(16):4851–4858. doi:10.1158/1078-0432.CCR-06-0134
15. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi:10.1016/j.febslet.2008.05.012
16. Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–1151. doi:10.1016/j.eururo.2013.12.003
17. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi:10.1016/j.canlet.2013.01.033
18. Sarfi M, Abbastabar M, Khalili E. Long noncoding RNAs biomarker-based cancer assessment. J Cell Physiol. 2019;234(10):16971–16986. doi:10.1002/jcp.28417
19. Xu L, Wu Y, Che X, et al. Cox-LASSO analysis reveals a Ten-lncRNA signature to predict outcomes in patients with high-grade serous ovarian cancer. DNA Cell Biol. 2019;38(12):1519–1528. doi:10.1089/dna.2019.4826
20. Liang W, Sun F. Identification of pivotal lncRNAs in papillary thyroid cancer using lncRNA-mRNA-miRNA ceRNA network analysis. PeerJ. 2019;7:e7441. doi:10.7717/peerj.7441
21. Wu B, Wang K, Fei J, et al. Novel threelncRNA signature predicts survival in patients with pancreatic cancer. Oncol Rep. 2018;40(6):3427–3437.
22. Zhou M, Guo M, He D, et al. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi:10.1186/s12967-015-0556-3
23. Fan Q, Liu B. Identification of a RNA-seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163–5172. doi:10.12659/MSM.902615
24. Zhang Y, Zhang L, Xu Y, Wu X, Zhou Y, Mo J. Immune-related long noncoding RNA signature for predicting survival and immune checkpoint blockade in hepatocellular carcinoma. J Cell Physiol. 2020.
25. Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. 2020;112(3):2640–2646. doi:10.1016/j.ygeno.2020.02.015
26. Sun J, Zhang Z, Bao S, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1):e000110. doi:10.1136/jitc-2019-000110
27. Wang W, Zhao Z, Yang F, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136(2):263–271. doi:10.1007/s11060-017-2667-6
28. Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol. 2018;55(5):3684–3697.
29. Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16(1):16. doi:10.1186/s12943-017-0580-4
30. Gu J-X, Zhang X, Miao R-C, et al. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J Gastroenterol. 2019;25(2):220–232. doi:10.3748/wjg.v25.i2.220
31. Tang J, Ren J, Cui Q, et al. A prognostic 10-lncRNA expression signature for predicting the risk of tumour recurrence in breast cancer patients. J Cell Mol Med. 2019;23(10):6775–6784. doi:10.1111/jcmm.14556
32. Fan Q, Liu B. Discovery of a novel six-long non-coding RNA signature predicting survival of colorectal cancer patients. J Cell Biochem. 2018;119(4):3574–3585. doi:10.1002/jcb.26548
33. Bao Z, Zhang W, Dong D. A potential prognostic lncRNA signature for predicting survival in patients with bladder urothelial carcinoma. Oncotarget. 2017;8(6):10485–10497. doi:10.18632/oncotarget.14441
34. Lian P, Wang Q, Zhao Y, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for bladder cancer. Aging (Albany NY). 2019;11(17):6930–6940. doi:10.18632/aging.102225
35. Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17(1):142. doi:10.1186/s12943-018-0893-y
36. Witkowski MT, Dolgalev I, Evensen NA, et al. Extensive remodeling of the immune microenvironment in B cell acute lymphoblastic leukemia. Cancer Cell. 2020;37(6):867–882 e812. doi:10.1016/j.ccell.2020.04.015
37. Varn FS, Wang Y, Mullins DW, Fiering S, Cheng C. Systematic pan-cancer analysis reveals immune cell interactions in the tumor microenvironment. Cancer Res. 2017;77(6):1271–1282. doi:10.1158/0008-5472.CAN-16-2490
38. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
39. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
40. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi:10.1038/ng1180
41. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
42. Riester M, Taylor JM, Feifer A, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18(5):1323–1333. doi:10.1158/1078-0432.CCR-11-2271
43. Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. doi:10.1158/1078-0432.CCR-12-0077-T
44. Zhou M, Zhang Z, Bao S, et al. Computational recognition of lncRNA signature of tumor-infiltrating B lymphocytes with potential implications in prognosis and immunotherapy of bladder cancer. Brief Bioinform. 2020.
45. Zhao D, Peng Q, Wang L, et al. Identification of a six-lncRNA signature based on a competing endogenous RNA network for predicting the risk of tumour recurrence in bladder cancer patients. J Cancer. 2020;11(1):108–120. doi:10.7150/jca.35801
46. Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63(2):234–241. doi:10.1016/j.eururo.2012.07.033
47. Woldu SL, Bagrodia A, Lotan Y. Guideline of guidelines: non-muscle-invasive bladder cancer. BJU Int. 2017;119(3):371–380. doi:10.1111/bju.13760
48. Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. doi:10.3727/096504016X14549667334007
49. Bin X, Hongjian Y, Xiping Z, Bo C, Shifeng Y, Binbin T. Research progresses in roles of LncRNA and its relationships with breast cancer. Cancer Cell Int. 2018;18:179. doi:10.1186/s12935-018-0674-0
50. Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9(7):1354–1366.
51. Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146(10):2901–2912
52. Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913.
53. Sun M, Nie FQ, Wang ZX, De W. Involvement of lncRNA dysregulation in gastric cancer. Histol Histopathol. 2016;31(1):33–39.
54. Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin Chim Acta. 2020;503:1–18. doi:10.1016/j.cca.2019.12.028
55. Wei C, Liang Q, Li X, et al. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem. 2019;120(9):14916–14927. doi:10.1002/jcb.28754
56. He RQ, Huang ZG, Li TY, et al. RNA-sequencing data reveal a prognostic four-lncRNA-based risk score for bladder urothelial carcinoma: an in silico update. Cell Physiol Biochem. 2018;50(4):1474–1495. doi:10.1159/000494647
57. Wang O, Yang F, Liu Y, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9(2):533–545.
58. Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. 2018;17(2):2743–2749.
59. Liang M, Pan Z, Yu F, Chen C. Long noncoding RNA SNHG12 suppresses esophageal squamous cell carcinoma progression through competing endogenous RNA networks. Clin Transl Oncol. 2020;22(10):1786–1795. doi:10.1007/s12094-020-02317-7
60. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36(1):11. doi:10.1186/s13046-016-0486-9
61. Song J, Wu X, Ma R, Miao L, Xiong L, Zhao W. Long noncoding RNA SNHG12 promotes cell proliferation and activates Wnt/beta-catenin signaling in prostate cancer through sponging microRNA-195. J Cell Biochem. 2019;120(8):13066–13075.
62. Tamang S, Acharya V, Roy D, et al. SNHG12: an LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. 2019;9:901.
63. Jiang B, Hailong S, Yuan J, et al. Identification of oncogenic long noncoding RNA SNHG12 and DUXAP8 in human bladder cancer through a comprehensive profiling analysis. Biomed Pharmacother. 2018;108:500–507. doi:10.1016/j.biopha.2018.09.025
64. Feng Z, Chen R, Huang N, Luo C. Long non-coding RNA ASMTL-AS1 inhibits tumor growth and glycolysis by regulating the miR-93-3p/miR-660/FOXO1 axis in papillary thyroid carcinoma. Life Sci. 2020;244:117298. doi:10.1016/j.lfs.2020.117298
65. Liang T, Wang B, Li J, Liu Y. LINC00922 accelerates the proliferation, migration and invasion of lung cancer via the miRNA-204/CXCR4 axis. Med Sci Monit. 2019;25:5075–5086. doi:10.12659/MSM.916327
66. Zhang H, Shan G, Song J, et al. Extracellular matrix-related genes play an important role in the progression of NMIBC to MIBC: a bioinformatics analysis study. Biosci Rep. 2020;40:5.
67. Zheng T, Wang J, Zhao Y, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi:10.1038/ncomms3996
68. Ziada AS, Lu MY, Ignas-Menzies J, et al. Mitochondrial DNA somatic mutation burden and heteroplasmy are associated with chronological age, smoking, and HIV infection. Aging Cell. 2019;18(6):e13018. doi:10.1111/acel.13018
69. Zhang Y. Understanding the gender disparity in bladder cancer risk: the impact of sex hormones and liver on bladder susceptibility to carcinogens. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2013;31(4):287–304. doi:10.1080/10590501.2013.844755
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.