Back to Journals » Cancer Management and Research » Volume 11
Expression of endoplasmic reticulum oxidoreductase 1-α in cholangiocarcinoma tissues and its effects on the proliferation and migration of cholangiocarcinoma cells
Authors Yan W, Wang X, Liu T, Chen L, Han L, Xu J, Jin G, Harada K, Lin Z, Ren X
Received 26 September 2018
Accepted for publication 13 June 2019
Published 19 July 2019 Volume 2019:11 Pages 6727—6739
DOI https://doi.org/10.2147/CMAR.S188746
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Wendi Yan,1,* Xue Wang,1,* Tesi Liu,1 Liyan Chen,2 Longzhe Han,3 Jing Xu,4 Guihua Jin,5 Kenichi Harada,6 Zhenhua Lin,1 Xiangshan Ren1,2
1Department of Pathology and Cancer Research Center, Yanbian University Medical College, Yanji 133002, People’s Republic of China; 2Key Laboratory of Natural Resources of Changbai Mountain & Functional Molecules (Yanbian University), Ministry of Education, Yanji 133002, People’s Republic of China; 3Department of Yanbian University Affiliated Hospital, Yanji 133002, People’s Republic of China; 4Department of Shanxi Medical University Medical College, Taiyuan 030001, People’s Republic of China; 5Department of Immunology and Pathogenic Biology, Yanbian University College of Medicine, Yanji 133002, People’s Republic of China; 6Department of Human Pathology, Kanazawa University Graduate School of Medicine, Kanazawa 920-8640, Japan
*These authors contributed equally to this work
Abstract: Endoplasmic reticulum oxidoreductase 1-α (ERO1A) is a kind of hypoxia-induced endoplasmic reticulum oxidase that regulates translation and folding of oxidized proteins. This study aimed to explore the clinicopathological significance of ERO1A and the effect on the biological behavior of cholangiocarcinoma (CCA) cells.
Methods: Immunohistochemical staining was used to detect the expression of ERO1A, carcinoembryonic antigen (CEA), and carbohydrate antigen 19–9 (CA19-9) in cholangiocarcinoma. Immunofluorescence staining was performed to detect the subcellular localization of ERO1A in CCA cells. The expression of ERO1A in CAA cells after depletion or overexpression was verified by Western blot assay. Then, the effect of ERO1A on proliferation in CCA cells was verified by MTT assay and colony formation assay. Wound healing assays and migration assays were performed to detect the effect of ERO1A on cell migration ability. Finally, we explored the role of ERO1A in EMT and Akt/mTOR signaling pathway.
Results: In this study, our data demonstrated that ERO1A, CEA, and CA19-9 were expressed in cholangiocarcinoma tissues, and the positive rates were 95%, 95%, and 55%, respectively. The high expression of ERO1A is associated with clinical stage and pathological stage of CCA. In vitro data indicate that deletion of ERO1A can inhibit the proliferation and migration of CCA cells and vice versa. In addition, ERO1A has been shown to be closely related to EMT and Akt/mTOR pathways.
Conclusion: In summary, we found that high expression of ERO1A is associated with poor prognosis in patients, and ERO1A can promote the proliferation and migration of CCA cells. In conclusion, ERO1A can be used as an independent biomarker for predicting the prognosis of CCA.
Keywords: ERO1A, cholangiocarcinoma, survival, prognosis, EMT
Introduction
Cholangiocarcinoma (CCA) is a highly aggressive malignant tumor that represents approximately 10% of all hepatobiliary malignancies.1 According to anatomical classification, it is roughly divided into intrahepatic cholangiocarcinoma (ICCA) and extrahepatic bile duct carcinoma (ECCA). Treatment options for CCA are limited, and overall rates of survival are low.2,3 Surgical treatment is the only curative option for all subtypes.4 However, the early diagnosis rate of tumors is extremely low, and most tumors are diagnosed in advanced stages. High recurrence rates at least 50–60% remain even after surgical resection5–7 There is no effective diagnostic method to improve the survival rate of patients. Therefore, to find a new biomarker has become an urgent problem.
EROlA is an oxidoreductase that is present in the endoplasmic reticulum, which regulates hypoxia-induced oxidative protein folding.8 This process is mediated by ERO1A and protein disulfide isomerase (PDI) catalysis, where ERO1A helps maintain the oxidative activity of PDI.9,10 ERO1A may be associated with apoptosis, infiltration, and endoplasmic reticulum stress (ERS)-induced metastasis.11,12 ERO1A is highly expressed in breast cancer, gastric cancer and is associated with the proliferation and migration of tumor cells.8,13 According to Kukita K, ERO1A is also highly expressed in colon cancer, and it can decrease tumorigenicity in vivo.14 Li G reported that ERO1A plays an important role in the induction of apoptosis.15 Therefore, ERO1A is highly expressed in various cancers and is closely related to tumor proliferation, migration, and apoptosis. Up to now, there is no relevant research reported on the interaction between ERO1A and cholangiocarcinoma.
In this study, we investigated the expression of ERO1A in cholangiocarcinoma, explored the relationship between ERO1A and clinicopathological features, and explored the effect of ERO1A expression on proliferation and migration of cholangiocarcinoma cells. This study can provide new ideas and effective molecular markers for the clinical treatment of cholangiocarcinoma.
Materials and methods
Tissue specimens
A total of 222 tissue microarrays and 20 cases of cholangiocarcinoma tissues of primary tumors in patients who were not treated before surgery, along with medical records, were collected from Outdo Biotech Co. Ltd. (Shanghai, China) between 2007 and 2016, including 124 cases of ICCA, 62 cases of ECCA, and 36 cases of adjacent tissue. There were 175 cases with survival information (including 104 cases of overall survival data and 71 cases of replase-free survival data). Survival time was counted from the date of surgery to the follow-up deadline or date of relapse. The study complied with the principles of the Declaration of Helsinki and was approved by the Human Ethics Committee and research ethics committees of Yanbian University Medical College in China.
Cell lines and cell culture
Human cholangiocarcinoma cell line CCKS1 was provided by the Department of Human Pathology at Kanazawa University,16 and it was approved by Ethics Committee of Kanazawa University. HuCCT1 and TFK1 were obtained from the Health Science Research Resources Bank (Osaka, Japan). Human cholangiocarcinoma cell lines RBE, QBC939 and human normal bile duct epithelial cells HIBepiC were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). HuCCT1, TFK1, RBE, QBC939, and HIBepiC were all cultured in RPMI-1640 (GIBCO, Gaithersburg, MD). The CCKS1 was cultured in Dulbecco Modified Eagle Medium (GIBCO, Gaithersburg, MD), supplemented with 10% fetal bovine serum (GIBCO, Gaithersburg, MD) and 1% penicillin and streptomycin mixture at 37°C in a humidified incubator with 5% CO2 atmosphere.
Immunohistochemistry staining
Immunohistochemistry (IHC) staining was routinely carried out using an immunohistochemistry kit (abcam, UK). The tissue sections were deparaffinized and rehydrated. The antigen was retrieved by sodium citrate buffer. Following endogenous peroxidase blocking and incubation in normal goat serum (Vector Lab, Burlingame, CA) for 20 mins, these sections were incubated with ERO1A (1:200, LSBio, USA), CEA (Use directly, zsbio, China), CA19-9 (Use directly, zsbio, China) overnight at 4°C and then with the peroxidase-conjugated secondary antibody (abcam, UK). Color development was performed with 3,3′-diaminobenzidine (DAB) and slides were counterstained with hematoxylin. Neutral resin sealing was performed. The results of immunohistochemical staining were observed under a microscope (Olympus Tokyo, Japan).
Immunohistochemistry score
Semi-quantitative IHC scores were interpreted according to a combination of staining intensity and rate of positive cancer cells. First, the scoring for staining intensity was: 0 point (negative), 1 point (weak), 2 points (moderate), and 3 points (intense). The percentage of positive tumor cells was assigned to five categories, which were 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. After multiplying the above two scores, the following scores 0–12 were obtained and further score was: 0 point, negative; 1–4 points, weakly positive; 6–8 points, moderately positive; and 9–12 points, strongly positive.17 Positive rate is the percentage of positive cases with any positive staining score, with 0 point defined as negative. Three pathologists (Xiangshan Ren, Zhenhua Lin, and Tiefeng Jin) examined and scored all tissue specimens.
The depletion and overexpression of ERO1A
Synthetic siRNA for ERO1A (Table 1) and nonsilencing siRNA (negative control, NC) were purchased from RiboBio (Guangzhou, China). The cells were incubated with Opti-MEM (Reduced Serum Medium, Gibco) containing premixed siRNA (50 nM) and 5 uL of Lipofectamine 3000 (Invitrogen, Carlsbad, CA), and were further incubated for 48 hrs. Lentiviral of overexpression ERO1A were constructed by VectorBuilder Inc (Table 1). The TFK1 cells were infected with 500 μL of complete medium containing 5 μg/mL of polybrene and 5 μL of lentiviral vector (titer=1×106TU/mL). At 8 hrs after infection, fresh medium was replaced and maintained for a further 72 hrs. ERO1A-transduced cells were then incubated with 1 μg/mL of puromycin in culture medium for 7 days to select stably transfected cells.
 |
Table 1 Interference sequences of ERO1A |
Western blot analysis
Total proteins were extracted from cultured cells using pierce T-PER protein extraction reagent (cwbiotech, China) and a sonicator (SONICS, USA). Protein quantification with BCA (Beyotime, Shanghai, China) Kit. Samples were denatured in sodium dodecyl sulfate (SDS) sample buffer. Total proteins were separated by loading 40 μg of total cell lysate onto SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane. The membrane was incubated with primary antibodies against ERO1A (1:1,000, LSBio, USA), actin (1:3,000, Abcam, USA), p-Akt (1:1,000, CST, USA), Akt (1:1,000, CST, USA), p-4EBP1 (1:1,000, CST, USA), 4EBP1 (1:1,000, CST, USA), p-S6 (1:1,000, CST, USA), S6 (1:1,000, CST, USA), E-cadherin (1:1,000, CST, USA), Vimentin (1:1,000, CST, USA), Snail (1:1,000, CST, USA), Slug (1:1,000, CST, USA), MMP-2 (1:1,000, CST, USA) and MMP-9 (1:1,000, CST, USA), and enzymatic signals were visualized using a chemiluminescence kit (Beyotime Biotechnology, China).
Immunofluorescence staining
The cells were seeded on 6-well culture plates at a density of 20,000 cells per well. The next day, the cells were fixed with 4% paraformaldehyde for 15 mins and permeabilized using 0.1% Triton X-100 for 10 mins (CWBIO, China). After blocking, the cells were incubated with a primary antibody against ERO1A (1:100, LSBio, USA) for overnight at 4°C. Alexa-488 (1:500, GeneCopoeia USA) was used as a secondary antibody, and nuclei were stained with 4′,6-diamino-2-phenylindole (Solarbio, China). The fluorescence signal was detected using a fluorescence microscope (Olympus Tokyo, Japan).
Proliferation assay
HuCCT1, QBC939, and TFK1 cells were seeded on 96-well plate at a density of 5,000 cells per well (n=6). After incubation for 48 hrs, 72 hrs, and 96 hrs at 37°C, the cells were incubated with 100 uL MTT (1 mg/mL, Amresco, USA) for 4 hrs. After adding 100 µL of DMSO (Amresco, USA), the value of absorbance was measured at a wavelength of 490 nm using a full-wavelength microplate reader (TECAN, Switzerland).
Colony formation assay
After cell silencing and lentiviral vector overexpression of ERO1A, cells in logarithmic growth phase were seeded in 6-well plates at a density of 500 cells per well. After the cells were incubated for 15 days, cells were then fixed with 4% paraformaldehyde for 15 mins and stained with 1% crystal violet (Solarbio, China). Statistical analysis was performed using Image J software.
Wound healing assay
Cells were seeded on 6-well plates, and wounds were created by 200 µL micropipette tip when cells were fused to 80%, replace the fresh medium. The effect of ERO1A on wound closure was observed by microscopy (Olympus, Japan). The results were analyzed by Image-J software.
Cell migration assay
Cell migration analysis was carried out using transwell inserts (Costar, Corning Incorporated, USA). Cells with medium containing 1% FBS were seeded in upper chamber. HuCCT1 and QBC939 cells were inoculated with 50,000 cells per pore, TFK1 cells with 100,000 cells per pore. The medium containing 10% FBS was placed in the lower chamber. HuCCT1 cells were incubated for 24 hrs, and QBC939 and TFK1 cells were incubated for 96 hrs. The cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of migrated cells was counted using microscope (KX4, Olympus, Japan).
Statistical analysis
Statistical analyses were primarily performed using SSPS17.0 (SSPS, Chicago, IL), and graphical interpretations were generated with GraphPad Prism 6 (GraphPad Software, San Diego, CA). The chi-square test was used to evaluate the correlation between ERO1A expression and clinicopathological features (statistics using percentages). The Kaplan–Meier method was used for the analysis of survival curves, and statistical significance was assessed using the Log-rank test. Cox proportional hazards regression model was used to examine univariate and multivariate hazard ratios for the study variables. Differences between groups were analyzed using a two-way ANOVA , and group comparisons for continuous data were done by a one-way ANOVA. P<0.05 was considered statistically significant.
Results
The expression of ERO1A in CCA
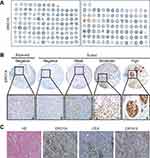
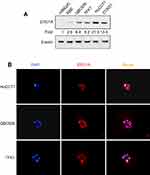
IHC staining was performed to detect the expression of ERO1A in cholangiocarcinoma (Figure 1A and B). The results showed that the positive staining ratio of ERO1A in cholangiocarcinoma was 84.9% (158/186), but in the adjacent non-tumor tissues was 0% (0/36) (Table 2). The expressions of ERO1A and CEA, CA19-9 were also compared, and the positive staining ratio was 95%, 95%, and 55% (Figure 1C, Table 3), respectively. Next, we examined the expression of ERO1A in cholangiocarcinoma cells. Western blot analysis revealed that protein levels of ERO1A were markedly upregulated in CCA cells (HuCCT1, CCKS1, QBC939, TFK1, and RBE) compared to normal bile duct epithelial cells (HIBepiC) (Figure 2A). Immunofluorescence staining showed that ERO1A was mainly expressed in cytoplasm (Figure 2B).
 |
Table 2 The ERO1A protein expression in cholangiocarcinoma |
 |
Table 3 The expression of ERO1A, CEA, and CA19-9 in cholangiocarcinoma |
Correlations between ERO1A expression and clinicopathological features in CCA
After confirming the high expression of ERO1A in cholangiocarcinoma, we further explored the correlation between ERO1A and clinicopathological features of patients with cholangiocarcinoma. Chi-square test showed that high expression of ERO1A was closely related to clinical stage (P<0.05) and pathological stage (P<0.05), but there was no significant correlation with age, gender, or tumor size (Table 4).
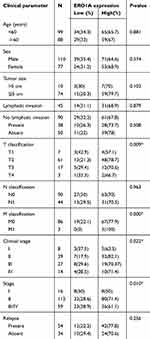 |
Table 4 Correlation between ERO1A expression and clinicopathologic features of cholangiocarcinoma cancer patients (statistics using percentages) |
High ERO1A expression was an independent biomarker for poor prognosis in CCA
To investigate the relationship between ERO1A expression and survival in patients with CCA, we analyzed overall survival (OS), recurrence-free survival (RFS), and median survival. The results showed that patients with high expression of ERO1A had a significantly shorter overall survival (P<0.05), indicating that ERO1A expression in CCA patients may have a significant impact on patient survival (Figure 3A–C). We further validated the above results by univariate and multivariate Cox regression analyses model, confirming the expression of ERO1A (P<0.05), tumor size (P<0.05), which is an independent risk factor for survival in patients with CCA and is significantly associated with poor prognosis in patients with CCA (Table 5).
 |
Table 5 Univariate and multivariate analysis of prognosis in patients with cholangiocarcinoma |
The effect of ERO1A on the proliferation and migration ability of CCA cells
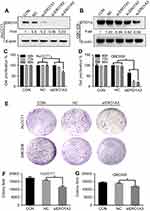
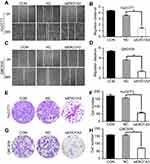
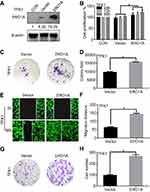
We further explored the effects of ERO1A on the biological behavior in CCA. Depleted or overexpressed ERO1A was verified by Western blotting (Figure 4A and B, Figure 6A). MTT assays (Figure 4C and D) and colony formation assays (Figure 4E and G) showed that ERO1A depletion can inhibit the proliferative capacity in HuCCT1 and QBC939 cells. As expected, the migration capacity of HuCCT1 and QBC939 was also reduced (Figure 5A–H). In contrast, the proliferation and migration abilities of TFK1 cells were significantly enhanced after overexpression of ERO1A (Figure 6B–H).
ERO1A regulates EMT progression in CCA
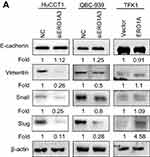
In view of the close relationship between EMT and tumor migration,18,19 and considering the effect of ERO1A on the metastasis of CCA cells, we examined the role of EMT in CCA by Western blot assays. The results showed that depletion of ERO1A caused a significant increase in E-cadherin, while Vimentin, Snail, and Slug were inhibited in HuCCT1 and QBC939 cells. As we speculated, the results were reversed in TFK1 cells (Figure 7A). In summary, ERO1A is involved in the regulation of EMT progression in cholangiocarcinoma cells.
ERO1A regulates the Akt/mTOR signaling pathways in CCA cells
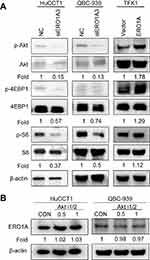
The Akt/mTOR signaling pathway plays an important role in cell growth, migration, and other processes. Disorders in signal transduction of this pathway may cause various diseases, including cancer.20 To further explore the mechanism by which ERO1A affects CCA cell proliferation and migration, we examined the expression of the Akt/mTOR signaling pathway. The results of Western blotting showed that the levels of p-Akt, p-4EBP1, and p-S6 were decreased after silencing ERO1A. Conversely, expression of p-Akt, p-4EBP1, and p-S6 was upregulated in TFK1 cells with ERO1A overexpression (Figure 8A). To further clarify this mechanism, we added the Akt signaling pathway inhibitor Akti-1/2, and the results showed that the change of ERO1A protein level was not obvious (Figure 8B). These results confirm that ERO1A is located upstream of the Akt signaling pathway. However, the more specific molecular mechanism of this result still requires further study.
Discussion
Studies have shown that ERO1A is highly expressed in other cancers, such as gastric cancer, breast cancer, and pancreatic ductal adenocarcinoma, and is associated with poor prognosis.21 This is consistent with the results of our study. Our research showed that ERO1A was highly expressed in CCA tissues, with a positive rate of 84.9%. The overall survival time of patients with high ERO1A expression was significantly shortened, and ERO1A was closely related to pathological and clinical stages. These results suggest that ERO1A can be used as an independent biomarker for predicting the prognosis of CCA patients.
Due to the lack of early symptoms or reliable tumor biomarkers, most CCA patients have reached an advanced stage when diagnosed. Reliable marker for the diagnosis of CCA is crucial for treatment and prognosis. At present, CEA and CA19-9 are commonly used in clinical diagnosis of cholangiocarcinoma. However, these markers are not specific because CEA and CA19-9 can be used as diagnostic markers for a variety of cancers and can also be found in some benign diseases such as cholangitis and intrahepatic bile duct stones.22,23 Loosen SH et al reported that CEA had more diagnostic values than CA19-9 in cholangiocarcinoma, which was consistent with the results of our study.24 Our results showed that the positive staining rate of ERO1A and CEA in 20 cases of cholangiocarcinoma was 95%, while the positive staining rate of CA19-9 was only 55%. The number of cases we tested was only 20, which did not fully reflect the situation, but it also suggested that it could be an effective molecular marker in cholangiocarcinoma. The specific mechanism remains to be further studied. With the advent of new technologies such as intraductal ultrasound in clinical screening, early CCA testing has become feasible, but these invasive tests may cause pain in patients.25 In recent years, tumor biomarkers have played an increasingly important role in diagnosis. According to an international multicenter study, very early cholangiocarcinoma is difficult to diagnose, especially when the tumor size ≦2 cm.26 Therefore, we tested the positive rate of ERO1A was 93% when the tumor volume was ≦2cm. However, whether ERO1A can be used clinically as an early diagnostic marker remains to be further studied.
According to the research by Zhang et al,27 ERO1A can regulate the proliferation, migration, and tumorigenesis of cervical cancer. Our hypothesis that ERO1A is involved in the proliferation and migration of cholangiocarcinoma. As we speculate, our results show that the proliferation and migration of cholangiocarcinoma cells were inhibited after ERO1A depleted, and the ability of proliferation and migration was enhanced when ERO1A was overexpressed.
Previous studies have reported that the expression of several tumor-associated factors is promoted by ERO1A, such as the master regulator for tumor metastasis.28 Therefore, we speculate whether ERO1A is associated with tumor metastasis and EMT. EMT also plays an important role in the development of these tumor phenotypes.29,30 Several studies have shown that EMT can accelerate the invasiveness of hepatocellular carcinoma cells.31–33 Our results validate our hypothesis that ERO1A can suppress the progression of EMT after depleted, and when it is overexpressed, it can accelerate the EMT process. These results further indicate that ERO1A may affect the migration of cholangiocarcinoma cells by regulating EMT.
The Akt/mTOR pathway regulates many cellular processes including proliferation and survival.34 Previous studies have shown that the proliferation of CCA cells was related to Akt/mTOR pathway.35 In this study, depletion of ERO1A in HuCCT1 and QBC-939 cells can reduce the expression of p-Akt, p-4EBP1, and p-S6, whereas the result of overexpression of ERO1A is the opposite. We are curious about the regulatory mechanism of ERO1A on this pathway, so we did further research. The results showed that the change in ERO1A protein level was not significant after the Akt pathway inhibitor was used. Then, we hypothesize that ERO1A is located upstream of the Akt signaling pathway. However, the specific mechanism of action remains to be further studied.
In conclusion, we indicated that ERO1A is aberrantly overexpressed in patients with cholangiocarcinoma and is closely related to its poor prognosis. As a cancer-promoting gene, ERO1A can accelerate the proliferation and migration of tumor cells. Therefore, ERO1A can be a promising biomarker and a potential therapeutic target for judging the prognosis of cholangiocarcinoma.
Highlights
- Endoplasmic reticulum oxidoreductase 1-α is overexpressed in cholangiocarcinoma and is a potential predictor of poor prognosis in patients with cholangiocarcinoma.
- Endoplasmic reticulum oxidoreductase 1-α is closely related to the proliferation and migration ability of cholangiocarcinoma cells.
Acknowledgments
This study was supported by three National Natural Science Funds of China (No. 31460303, No. 81660609, No. 81460255), and Jilin Provincial Science and Technology Department Project (No. 20180101007JC). We thank James P. Mahaffey, PhD, from Liwen Bianji, Edanz Editing China, for editing the English text of a draft of this manuscript.
Disclosure
Prof. Dr Zhenhua Lin reports grants from Key Laboratory of the Science and Technology Department of Jilin Province, during the conduct of the study; outside the submitted work. The authors report no other conflicts of interest in this work.
References
1. Sapisochín G, Sevilla EFD, Echeverri J, et al. Liver transplantation for cholangiocarcinoma current status and new insights J. World J Hepatol. 2015;7(22):2396–2403. doi:10.4254/wjh.v7.i22.2396
2. Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–1229. doi:10.1053/j.gastro.2013.10.013
3. Cardinale V, Semeraro R, Torrice A, et al. Intra-hepatic and extra-hepatic cholangiocarcinoma: new insight into epidemiology and risk factors. World J Gastrointest Oncol. 2010;2(11):407. doi:10.4251/wjgo.v2.i11.407
4. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi:10.1016/S0140-6736(13)61612-8
5. Nathan H, Pawlik TM, Wolfgang CL, Cameron JL, Schulick RD, Schulick RD. Trends in survival after surgery for cholangiocarcinoma: a 30-year population-based SEER database analysis. J Gastrointest Surg. 2007;11:1488–1496. doi:10.1007/s11605-007-0282-0
6. Mavros MN, Economopoulos KP, Alexiou VG, Pawlik TM. Treatment and prognosis for patients with intrahepatic cholangiocarcinoma: systematic review and meta-analysis. JAMA Surg. 2014;149:565–574. doi:10.1001/jamasurg.2013.5137
7. Kerr SE, Barr Fritcher EG, Campion MB, et al. Biliary dysplasia in primary sclerosing cholangitis harbors cytogenetic abnormalities similar to cholangiocarcinoma. Hum Pathol. 2014;45(9):1797–1804. doi:10.1016/j.humpath.2014.05.008
8. Seol SY, Kim C, Lim JY, et al. Overexpression of endoplasmic reticulum oxidoreductin 1-α (ERO1A) is associated with poor prognosis of gastric cancer. Cancer Res Treat. 2016;48(4):1196–1209. doi:10.4143/crt.2015.189
9. Masui S, Vavassori S, Fagioli C, Sitia R, Inaba K. Molecular bases of cyclic and specific disulfide interchange between human ERO1α Protein and Protein-disulfide Isomerase (PDI). J Biol Chem. 2011;286(18):16261. doi:10.1074/jbc.M111.231357
10. Araki, Kazutaka, Kazuhiro Nagata. Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J Biol Chem. 2011;286(37):32705–32712. doi:10.1074/jbc.M111.227181
11. Periyasamy P, Guo ML, Buch S, et al. Cocaine induces astro-cytosis through ER stress-mediated activation of autophagy. Autophagy. 2016;12:1310–1329. doi:10.1080/15548627.2016.1183844
12. Zhang ZZ, Yuan K, Yue HT, et al. Identification and functional characterization of an endoplasmic reticulum oxidoreductin 1-α gene in litopenaeus vannamei. Dev Comp Immunol. 2016;57:10–19. doi:10.1016/j.dci.2015.11.013
13. Kutomi G, Tamura Y, Tanaka T, et al. Human endoplasmic reticulum oxidoreductin 1-a is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013;104:1091–1096. doi:10.1111/cas.12177
14. Takei N, Yoneda A, Sakai-Sawada K, Kosaka M, Minomi K, Tamura Y. Hypoxia-inducible ERO1α promotes cancer progression through modulation of integrin-β1 modification and signalling in HCT116 colorectal cancer cells. Sci Rep. 2017;7(1):9389. doi:10.1038/s41598-017-09976-7
15. Li G, Mongillo M, Chin KT, et al. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186(6):783–792. doi:10.1083/jcb.200904060
16. Sugawara H, Yasoshima M, Katayanagi K, et al. Relationship between interleukin-6 and proliferation and differentiation in cholangiocarcinoma. Histopathology. 1998;33:145–153. doi:10.1046/j.1365-2559.1998.00445.x
17. Jin T, Kim HS, Choi SK, et al. microRNA-200c 141 upregulates serpinB2 to promote breast cancer cell metastasis and reduce patient survival. Oncotarget. 2017;8(20):32769–32782. doi:10.18632/oncotarget.15680
18. Brabletz T. To differentiate or not–routes towards metastasis. Nat Rev Cancer. 2012;12(6):425–436. doi:10.1038/nrc3265
19. Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi:10.1016/j.ceb.2016.06.002
20. Baoyu L, Yang J, Lu Z, Liu B, Liu F. A study on the mechanism of rapamycin mediating the sensitivity of pancreatic cancer cells to cisplatin through PI3K/AKT/mTOR signaling pathway. J Buon. 2019;24(2):739–745.
21. Li H, Wang X, Fang Y, et al. Integrated expression profiles analysis reveals novel predictive biomarker in pancreatic ductal adenocarcinoma. Oncotarget. 2017;8(32):52571–52583.
22. Chen CY, Shiesh SC, Tsao HC, Lin X-Z. The assessment of biliary CA 125, CA 19-9 and CEA in diagnosing cholangiocarcinoma–the influence of sampling time and hepatolithiasis. Hepatogastroenterology. 2002;49:616–620.
23. Maestranzi S, Przemioslo R, Mitchell H, Sherwood RA. The effect of benign and malignant liver disease on the tumour markers CA19-9 and CEA. Ann Clin Biochem. 1998;35(Pt 1):99–103. doi:10.1177/000456329803500113
24. Loosen SH, Roderburg C, Kauertz KL, et al. CEA but not CA19-9 is an independent prognostic factor in patients undergoing resection of cholangiocarcinoma. Sci Rep. 2017;7(1):16975. doi:10.1038/s41598-017-17175-7
25. Sapisochin G, Facciuto M, Rubbia-Brandt L, et al. Liver transplantation for “very early” intrahepatic cholangiocarcinoma: international retrospective study supporting a prospective assessment. Hepatology. 2016;64:1178–1188. doi:10.1002/hep.28744
26. Rizvi S, Gores GJ. Emerging molecular therapeutic targets for cholangiocarcinoma. J Hepatol. 2017;67(3):632–644. doi:10.1016/j.jhep.2017.03.026
27. Zhang Y, Li T, Zhang L, et al. Targeting the functional interplay between endoplasmic reticulum oxidoreductin-1α and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine. 2019;41:408–419. doi:10.1016/j.ebiom.2019.02.041
28. Tanaka T, Kutomi G, Kajiwara T. Cancer-associated oxidoreductase ERO1-α drives the production of VEGF via oxidative protein folding and regulating the mRNA level. Br J Cancer. 2016;114(11):1227–1234.
29. Ke A, Shi G, Zhou J, et al. CD151 amplifies signaling by integrin α6β1 to PI3K and induces the epithelial–mesenchymal transition in HCC cells. Gastroenterology. 2011;140(5):1629–1641. doi:10.1053/j.gastro.2011.02.008
30. Ding W, You H, Dang H, et al. Epithelial-to-mesenchymal transition of murine liver tumor cells promotes invasion. Hepatology. 2010;52(3):945–953. doi:10.1002/hep.23748
31. Joseph JP, Harishankar MK, Pillai AA, Devi A. Hypoxia induced EMT: a review on the mechanism of tumor progression and metastasis in OSCC. Oral Oncol. 2018;80(May 2018):23. doi:10.1016/j.oraloncology.2018.03.004
32. Sun S, Ning X, Zhang Y, et al. Hypoxia-inducible factor-1α induces twist expression in tubular epithelial cells subjected to hypoxia, leading to epithelial-to-mesenchymal transition. Kidney Int. 2009;75(12):1278–1287. doi:10.1038/ki.2009.62
33. Liu Z, Tu K, Wang Y, et al. Hypoxia accelerates aggressiveness of hepatocellular carcinoma cells involving oxidative stress, epithelial-mesenchymal transition and non-canonical hedgehog signaling. Cell Physiol Biochem. 2017;44(5):1856. doi:10.1159/000485821
34. Lin HL, Yang MH, Wu CW, et al. 2-Methoxyestradiol attenuates phosphatidylinositol 3-kinase/Akt pathway-mediated metastasis of gastric cancer. Int J Cancer. 2007;121(11):2547–2555. doi:10.1002/ijc.22861
35. Ke F, Wang Z, Song X, et al. Cryptotanshinone induces cell cycle arrest and apoptosis through the JAK2/STAT3 and PI3K/Akt/NFκB pathways in cholangiocarcinoma cells. Drug Des Devel Ther. 2017;11:1753–1766. doi:10.2147/DDDT.S132488
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.