Back to Journals » OncoTargets and Therapy » Volume 12
Expression level and clinical significance of SNHG1 in human cancers: a meta-analysis
Authors Yu Y, Yang J, Yang S, Li Q, Zhang M, Wang L , Ji G, Miao L
Received 21 August 2018
Accepted for publication 11 March 2019
Published 24 April 2019 Volume 2019:12 Pages 3119—3127
DOI https://doi.org/10.2147/OTT.S184803
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Yang Yu,1 Jian Yang,2 Shengquan Yang,3 Quanpeng Li,1 Mingjiong Zhang,1 Lijuan Wang,4 Guozhong Ji,1 Lin Miao1
1Medical Centre for Digestive Diseases, Second Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu Province, People’s Republic of China; 2Department of Urology, Second Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu Province, People’s Republic of China; 3Department of Orthopaedics, The No. 1 People’s Hospital of Yancheng, Yancheng, Jiangsu Province, People’s Republic of China; 4Department of Geriatrics, The Second Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu Province, People’s Republic of China
Background: As reported by numerous research studies, the expression levels of SNHG1 (small nucleolar RNA host gene 1) are increased in different kinds of tumours, revealing that SNHG1 is likely to perform a crucial function in cancer prevalence and progression. Moreover, a mounting degree of evidence suggests that increased SNHG1 expression also has an association with poor medical outcomes among cancer patients.
Materials and methods: Collection of qualifying research studies was performed through the retrieval of keywords in PubMed and Web of Science, up to March 20, 2018. This quantitative meta-analysis was carried out using Stata SE12.0 software and aimed at exploring the connection between the expression level of SNHG1 and clinicopathology.
Results: Ten research studies, involving an aggregate of 715 patients, met the inclusion criteria. As suggested by the findings of the current meta-analysis, with regard to prognosis, the patients with high expression of SNHG1 had poorer overall survival (OS) (HR =3.36, 95% CI: 2.42, 4.67) and, with regard to their clinicopathology, increased SNHG1 was associated with advanced TNM stage (RR =1.88, 95% CI: 1.58, 2.24), poorly differentiated histological grade (RR =1.38, 95% CI:1.09, 1.76), and positive lymph node metastasis (RR =1.80, 95% CI: 1.42, 2.29).
Conclusion: As revealed by this meta-analysis, elevated SNHG1 expression is typical in various types of cancer. In addition, elevated SNHG1 expression is likely to function as an advanced predictive element of poor prognosis and lymph node metastasis in various cancer types. Nonetheless, to date, it remains essential to carry out larger-size and better-designed research studies for the confirmation of our findings.
Keywords: SNHG1, cancer, prognosis, meta-analysis
Introduction
Long noncoding RNAs (lncRNAs) represent a large family of RNAs without protein-coding capability that are characterized by a length of more than 200 nucleotides and the lack of an identifiable open reading frame (ORF).1–6 As revealed by the research to date, upregulation of some lncRNAs is evident in several cancer tissues and cell lines, in comparison to the tissues surrounding the cancer and normal cell lines, respectively. Additionally, some of these lncRNAs have pro-oncogenic potential; conversely, some others are reported to have low expression levels and tumour-suppressive functions.7–10
Among the lncRNAs, the small nucleolar RNA host gene 1 (SNHG1; also known as UHG, U22HG, lncRNA16, LINC00057, and NCRNA00057) has garnered our attention. Specification of SNHG1 mechanisms extends our understanding of invasive pathophysiology. Additionally, overexpression of SNHG1 is reported to be a predictor of oncogenesis in patients with many kinds of cancer, including oesophageal squamous cell carcinoma,11 lung squamous cell carcinoma,12 hepatocellular carcinoma,13,14 colorectal cancer15,16 and gastric cancer.17
As revealed by the growing number of research studies, SNHG1 is likely to perform the function of a diagnostic and a prognostic biomarker with regard to the cancers stated above. For the purpose of validating its clinical relevance as a biomarker or therapeutic target, it was deemed quite necessary to investigate whether SNHG1 expression level is associated with pathological features. The current research study is aimed at carrying out a meta-analysis of this association in human cancers.
Materials and methods
Literature search strategies
Independent identification of the relevant literature was performed by two scholars with the use of both PubMed and Web of Science. The relevant literature reported the connection between SNHG1 expression level and pathological attributes in human cancers. The literature search strategy included a combination of keywords (“SNHG1”, “LINC00057”, “lncRNA16”, “cancer or carcinoma or tumour or neoplasm”, and “pathology”). In addition, the references of attained literature were also examined for the identification of additional relevant studies.
Inclusion and exclusion criteria
The research studies involved in this analysis were required to satisfy the inclusion criteria listed here: 1) reported expression levels of SNHG1, as determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR); 2) put forward the decision; 3) segregated the patients into high and low expression groups with the help of definite criteria for SNHG1 expression levels; 4) reported data associated with the clinicopathological attributes of the patients, and at a minimum, one of the following pathological features: TNM stage, histological grade, lymph node metastasis, distant metastasis, and overall survival information; and 5) utilized a case-control or cohort study design.
Research studies were not included in the analysis if they met any of the exclusion criteria listed here: 1) stated recurring research reports or studies that included patients who were reported in a former research study; 2) provided insufficient statements of the data; 3) employed nonhuman specimens; 4) were reviews, together with letters, unpublished data, and commentaries; and 5) were reports that were not published in the English language.
Assessment of the quality of the research studies was performed by two scholars by going through the title, abstract, and complete text of each report while referencing the inclusion and exclusion criteria.
Literature screening and data extraction
Independent collection of the data was performed by two investigators (Yang Yu and Jian Yang), in accordance with both the inclusion and exclusion criteria, and after resolving any conflicts with the help of a consensus or talks with a third scholar (Shengquan Yang) prior to the performance of the analysis. Data extraction from the literature included the following: first author, publication year, country of data source, kind of cancer, number of patients placed in both the high and low SNHG1 expression cohorts, the SNHG1 expression level identification methodology, and the cut-off approximations for SNHG1 expression levels.
Quality assessment
Assessment of the quality of the involved research studies was performed with the help of the Newcastle-Ottawa Scale standard that assessed selection (four points), comparability (two points), and outcome (three points) and had a score ranging between 0 and 9. Each of the qualifying studies was scored in Table 1, with a higher score suggesting better methodological quality.
 | Table 1 Characteristics of the included studies |
Statistical analysis
Cochran’s Q and Chi-square-based I2 tests were used for the determination of the heterogeneity among the involved research studies. Homogeneity tests were carried out using a significance level of α =0.1. P-values <0.1 were regarded as being statistically significant, whereas I2 values >50% implied heterogeneity among the research studies. Analysis of the homogeneous data was carried out with the use of a fixed effects framework; otherwise, a random effects framework was used for the analysis. Statistical analyses, together with the assessment of publication bias and Begg’s methodology, was carried out using StataSE 12.0 (Stata Corp LP, College Station, Texas, USA).
Results
Data selection and characteristics
Ten research studies that involved an aggregate of 715 patients showed agreement with the inclusion criteria. Each and every research study originated from China; two were studies of colorectal cancer, two were studies of hepatocellular carcinoma, two were studies of lung cancer, two were studies of osteosarcoma, and the remaining two were studies of oesophageal squamous cell carcinoma and gastric cancer. QRT-PCR was used for detecting SNHG1, on the basis of which, categorization of the patients into groups of high and low SNHG1 expression was performed. The mean and median were employed as the cut-off values for estimating for the SNHG1 expression level. A summary of the attributes of the involved research studies is provided in Table 1, whereas the flowchart of the study search, together with the selection process, is presented in Figure 1.
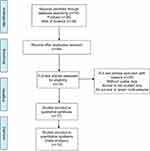 | Figure 1 Flowchart of selecting studies for inclusion. |
Association between SNHG1 expression and pathological features
TNM stage
Eight research studies reported a link between SNHG1 expression and TNM stage (III/IV versus I/II). There was no statistically significant (P>0.05, I2=0.00%) heterogeneity among the research studies; accordingly, the fixed-effects framework was applied for the calculation of the accumulated pooled RR, together with its 95% CI, which reached statistical significance [RR =1.88, 95% CI (1.58, 2.24), P<0.001] (Figure 2, Table 2). This suggests that a high SNHG1 expression level has a link with advanced TNM phase.
 | Table 2 Meta analysis results for the association of over-expressed SNHG1 with clinicopathological parameters |
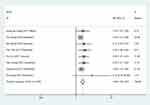 | Figure 2 Forest plot for the association between SNHG1 expression and TNM stage in human cancers. |
Histological grade
Reports from an aggregate of 4 research studies revealed the association between SNHG1 expression and histological grade. Statistically significant (P>0.05, I2=0.00%) heterogeneity among the research studies was not observed; accordingly, the fixed-effects framework was applied for the calculation of the accumulated RR, together with its 95% CI, which exhibited a statistically significant difference [RR =1.38, 95% CI (1.09, 1.76), P<0.01] (Figure 3, Table 2). This suggested that high SNHG1 expression was associated with a higher risk of poorly differentiated histological grade.
 | Figure 3 Forest plot for the association between SNHG1 expression and histological grade in human cancers. |
Lymph node metastasis
Reports from an aggregate of 5 research studies suggested a connection between SNHG1 expression and lymph node metastasis. Statistically significant (P>0.05, I=0.00%) heterogeneity was not observed among the studies; accordingly, the fixed-effects framework was applied for the calculation of the accumulated RR, together with its 95% CI, which reached statistical significance [RR =1.8, 95% CI (1.42, 2.29), P<0.001] (Figure 4, Table 2). This association illustrates the fact that the cohort with the high SNHG1 expression level exhibited a higher risk of lymph node metastasis compared with the cohort with the low SNHG1 expression levels.
 | Figure 4 Forest plot for the association between SNHG1 expression and lymph node metastasis in human cancers. |
Distant metastasis
Reports from 3 research studies revealed a link between SNHG1 expression level and distant metastasis. Statistically significant (P>0.05, I=0.00%) heterogeneity among the research studies was not observed; accordingly, the fixed-effects framework was applied for the calculation of the accumulated RR, together with its 95% CI, which did not reach statistical significance [RR =1.29, 95% CI (0.80, 2.08), P>0.05] (Figure 5, Table 2). This result highlights that SNHG1 expression levels have no correlation with distant metastasis. The reasons behind the inexistence of any correlation could include that the number of patients registered in some of the research works was comparatively smaller; moreover, not every kind of cancer was studied, and no consensus on the cut-off for making a distinction between a high or low SNHG1 expression level was observed. As such, future studies comprising larger samples of patients are going to be needed; moreover, the cut-off value for making a distinction between high or low SNHG1 expression level requires consistency as well.
 | Figure 5 Forest plot for the association between SNHG1 expression and distant metastasis in human cancers. |
Association between SNHG1 expression and survival in different types of cancers
In total, 5 research studies comprising 352 patients were employed for the assessment of the impact of SNHG1 overexpression on OS in various cancers (Table 3). Moreover, it was highlighted that augmented SNHG1 expression forecasted a weak performance for OS in the involved cancer types [pooled HR=3.36, 95% CI (2.42, 4.67), P<0.001], with heterogeneity (I2=57.3%, P>0.05). In addition, a subgroup analysis was also performed on the basis of the aggregate number of each study concerned. A substantial connection between SNHG1 overexpression and poor OS in the research studies with aggregate numbers below 80 patients was noted [pooled HR =2.55, 95% CI (1.52, 4.26)) (Figure 6). In comparison with the low SNHG1 expression cohort, the high SNHG1 expression group exhibited a statistically significant decline in OS, in addition to being correlated with worse survival.
 | Table 3 Characteristics of the overall survival of the included studies |
 | Figure 6 Meta-analysis for the pooled HRs of overall survival in patients with various cancers. |
Assessment of publication bias
Due to the small number of research studies included, analysis of publication bias was not possible for TNM phase, histological grade, lymph node metastasis, or distant metastasis.
Discussion
To date, deregulation of lncRNAs has been observed in numerous human cancers. Additionally, deregulation of lncRNAs has been linked to cancer proliferation by acting as a regulator in alternative splicing and translation, by promoting steadiness of the host mRNAs with the help of post-transcriptional phenomena, or by acting as the scaffolding or instructions for regulating protein-protein or protein-DNA interactions.18,19
The current meta-analysis was aimed at investigating the link between SNHG1 expression levels and pathological attributes observed in human cancers. In total, 715 patients from 10 research studies were eventually included. The fixed-effects framework was applied for the assessment of TNM stage and histological grade, lymph node metastasis, and distant metastasis. Consequently, the cohort with the high SNHG1 expression level exhibited a higher risk of advanced TNM stage, poorly differentiated grade and lymph node metastasis compared to the low SNHG1 expression level group. In addition, in regard to prognosis, the patients with high expression of SNHG1 experienced relatively short overall survival.
Nonetheless, this research study has some limitations: (1) each and every involved research study originated from China and no patients from any other country were included; (2) the number of patients registered in some of the research studies was comparatively smaller and not all the cancer types were studied; (3) no consensus was reached regarding the cut-off level for making a distinction between a high or low SNHG1 expression level; (4) no cohort studies observed met the inclusion criteria. High-quality studies with large sample sizes are necessary for the confirmation of these results.
To summarize, with regard to the clinicopathology, high expression levels of SNHG1 had a close association with advanced TNM stage, poorly differentiated grade, and lymph node metastasis. In addition, with regards to prognosis, the patients having a high expression level of SNHG1 experienced relatively poor overall survival (OS). Notably, SNHG1 is capable of acting as a biomarker of poor prognosis for patients with cancer.
Conclusion
This meta-analysis discovered that augmented SNHG1 expression is common in a number of different kinds of cancer and has a likelihood of acting as an innovative predictive element of poor prognosis and lymph node metastasis in various cancers. Nonetheless, it is still deemed essential to carry out research studies with a larger sample size, and with an improved design, for the confirmation of our findings.
Acknowledgments
This research work received the support from the Project of Standard Diagnosis and Treatment of Key Disease of Jiangsu Province (BE2015722), the Project of the Peak of the Six Talents of Jiangsu Province (WSN-018) and the Scientific Research Foundation for Health of Jiangsu Province (H201408).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi:10.1016/j.cell.2015.12.017
2. Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3ʹ UTRs. RNA. 2012;18:825–843. doi:10.1261/rna.029520.111
3. Zhao J, Song X, Wang K. lncScore: alignment-free identification of long noncoding RNA from assembled novel transcripts. Sci Rep. 2016;6:34838. doi:10.1038/srep34838
4. Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. Rna-A Publ Rna Soc. 2010;16:1478–1487. doi:10.1261/rna.1951310
5. Saha A, Bhattacharya S, Bhattacharya A. Serum stress responsive gene EhslncRNA of Entamoeba histolytica is a novel long noncoding RNA. Sci Rep. 2016;6:27476. doi:10.1038/srep27476
6. Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi:10.1146/annurev-genet-120213-092323
7. Li Z, Yu X, Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol. 2016;37:5657–5661. doi:10.1007/s13277-016-4808-5
8. Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi:10.1186/1471-2407-14-319
9. Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi:10.1158/0008-5472.CAN-12-2850
10. Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi:10.18632/oncotarget.1902
11. Zhang Y, Jin X, Wang Z, Zhang X, Liu S, Liu G. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2017;21:89–96. doi:10.3233/CBM-170286
12. Zhang HY, Yang W, Zheng FS, Wang YB, Lu JB. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–658. doi:10.1016/j.biopha.2017.03.104
13. Zhang H, Zhou D, Ying M, et al. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–4829.
14. Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi:10.1016/j.biopha.2016.02.036
15. Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. 2018;9:2128–2139. doi:10.18632/oncotarget.23255
16. Zhu Y, Li B, Liu Z, et al. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:111715–111727. doi:10.18632/oncotarget.22903
17. Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–931. doi:10.1016/j.bbrc.2017.07.137
18. Simon MD, Pinter SF, Fang R, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi:10.1038/nature12719
19. Engreitz JM, Pandya-Jones A, McDonel P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi:10.1126/science.1237973
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
