Back to Journals » International Journal of General Medicine » Volume 14
Expression, Clinical Significance, and Prospective Pathway Signaling of miR-501-3p in Ovarian Cancer Based on Database and Informatics Analysis
Authors Chen B , Jin X , Wang H, Zhou Q, Li G, Lu X
Received 7 July 2021
Accepted for publication 24 August 2021
Published 3 September 2021 Volume 2021:14 Pages 5193—5201
DOI https://doi.org/10.2147/IJGM.S327673
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Buze Chen,1,2,* Xin Jin,3,* Haihong Wang,1 Qingmei Zhou,1 Guilin Li,3 Xiaoyuan Lu1
1Department of Gynecology, The Affiliated Hospital of Xuzhou Medical University, Xuzhou, Jiangsu, 221000, People’s Republic of China; 2Xuzhou Medical University, Xuzhou, Jiangsu, 221000, People’s Republic of China; 3Department of Gynecology, Maternal and Child Health Care Hospital Affiliated to Xuzhou Medical University, Xuzhou, Jiangsu, 221000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaoyuan Lu
Department of Gynecology, The Affiliated Hospital of Xuzhou Medical University, No. 99 West Huaihai Road, Quanshan District, Xuzhou, 221000, Jiangsu, People’s Republic of China
Tel +86-180 52268119
Email [email protected]
Guilin Li
Department of Gynecology, Maternal and Child Health Care Hospital Affiliated to Xuzhou Medical University, No. 46 Heping Road, Yunlong District, Xuzhou, Jiangsu, 221000, People’s Republic of China
Tel +86-18936372957
Email [email protected]
Background: The present study aims to explore the expression, clinical significance, and prospective pathway signaling of miR-501-3p in ovarian cancer (OC) based on database and informatics analysis.
Methods: Kruskal–Wallis test, Wilcoxon sign-rank test, and logistic regression were used to evaluate the relationship between clinical features and miR-501-3p expression. Kaplan–Meier survival curve analysis was used to explore the relationship between miR-501-3p expression and the prognosis of OC patients. The miRNA targets were obtained from databases TargetScan, miRanda, TarBase, miRTarBase, miR2Disease, miRecords, and miRWalk. GO and KEGG analyses were used to analyze the significant involvement of miR-501-3p target genes in function.
Results: The low miR-501-3p expression in OC was significantly associated with histologic grade (P=0.015). Low miR-501-3p expression predicted a poorer overall survival (HR: 0.77; 95% CI: 0.61– 0.96; P=0.02) and disease-specific survival (HR: 0.77; 95% CI: 0.61– 0.99; P=0.038). GO and KEGG analyses demonstrated that miR-501-3p might participate in the development of OC by pathways including one carbon pool by folate, protein digestion and absorption, cell cycle, kaposi sarcoma-associated herpesvirus infection, and viral carcinogenesis.
Conclusions: Low miR-501-3p expression is significantly associated with poor survival in OC patients. It may be a promising prognostic biomarker for OC patients.
Keywords: microRNA-501-3p, ovarian cancer, prognosis, the cancer genome atlas, gene expression
Introduction
Ovarian cancer (OC) is one of the world’s deadliest gynaecological malignancies, causing over 140,000 deaths each year.1 Worldwide, 239,000 new patients are affected by ovarian cancer each year and 152,000 OC patients die.2 To date, the combination of surgery and platinum/paclitaxel chemotherapy has been used as the primary treatment for OC.3 The five-year survival rate for patients with early-stage OC is as high as 92%.4 However, despite the tremendous advances in various therapies over the past decades, only 19% of ovarian cancer patients are diagnosed early due to the lack of typical clinical signs and symptoms, the rapid rate of tumor progression, and the low 5-year survival rate.5 Therefore, the rapid search for potential biomarkers and therapeutic targets is of great significance to improve the prognosis of OC patients.
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression levels by binding to the 3ʹ-untranslated region (UTR) of their target mRNA.6 Aberrant expression of miR-501-3p is associated with the diagnosis or prognosis of certain cancers. MicroRNAs play a critical role in the development and progression of OC.7,8 MiR-501-3p can be used as a therapeutic diagnostic biomarker for non-small cell lung cancer (NSCLC) patients.9 MiR-501-3p is a new target for the molecular treatment of pancreatic ductal adenocarcinoma (PDAC).4 miR-501-3p can be used as a potential prognostic predictor and therapeutic tool for hepatocellular carcinoma (HCC).10 miR-501-3p is a new therapeutic target for osteosarcoma (OS).11 However, the role of miR-501-3p in OC is not yet clear.
In this study, we explored the expression of miR-501-3p in OC tumor tissues and normal ovarian tissues, the relationship between miR-501-3p expression and clinical characteristics, the relationship between miR-501-3p and patient prognosis, and the possible regulatory network of miR-501-3p mediating ovarian carcinogenesis and progression. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was used to determine the significant involvement of miR-501-3p target genes in function. This study provides a promising diagnostic and prognostic marker for patients with OC.
Patients and Methods
Differential Expression of miR-501-3p
Samples of tumor tissues and normal ovarian tissues from 10 HGSOC patients were collected at the Department of Obstetrics and Gynaecology of the Affiliated Hospital of Xuzhou Medical University. This study was approved by the ethics committee of Xuzhou Medical University Affiliated Hospital. All patients signed a written informed consent form. As shown in Table S1, the clinical characteristics of ten HGSOC patients were listed.
The miR-501-3p levels in 10 HGSOC tissue samples were identified by qRT-PCR. The detailed steps were carried out according to the literature.12 The primer sequences used were shown in Table S2.
Clinical Information
Software: R (version 3.6.3) (statistical analysis and visualization). R package: Basic R package. Molecule: hsa-miR-501-3p [MIMAT0004774]. Subgroups: Median. Data: miRNAseq data and clinical data from level 3 BCGSC miRNA Profiling in TCGA (https://portal.gdc.cancer.gov/) OC (ovarian cancer) project. Data filtering: Data on clinical information retained. Data transformation: miRNAseq data in RPM (Reads per Million mapped reads) format are grouped according to molecular expression.
The Correlation Between miR-501-3p Expression and Clinical Information
Software: R (version 3.6.3). R package: Basic R package and ggplot2 (for visualization). Molecule: hsa-miR-501-3p. Subgroup: median. Clinical variables: and histologic grade and venous invasion. Data: miRNAseq data and clinical data from level 3 BCGSC miRNA Profiling in the TCGA OC project. Data filtering: data with clinical information retained. Data transformation: miRNAseq data in RPM format grouped according to molecular expression.
The Prognosis Value of miR-501-3p Expression in OC Patients
Software: R (version 3.6.3). R packages: survminer package (for visualization), survival package (for statistical analysis of survival data). Molecule: hsa-miR-501-3p. Subgroup: Minimal p-value subgroup. Prognosis type: OS and DSS. Data: miRNAseq data from level 3 BCGSC miRNA Profiling and clinical data from the TCGA OC project. Data filtering: Retain data with clinical information. Data transformation: miRNAseq data in RPM format are grouped according to molecular expression and then analyzed.
Prediction of Putative miR-501-3p
The targets of miR-501-3p were predicted by databases including TargetScan, miRanda, TarBase, miRTarBase, miR2Disease, miRecords, and miRWalk.13–16 The differentially expressed genes (DEGs) of OC were obtained from GSE14407 by GEO2R. The common genes predicted by databases and OC down-regulated genes were analyzed on the basis of a Venn diagram.
GO and KEGG Analysis of miR-501-3p
Enrichment Analysis
Software: R (version 3.6.3). R packages: mainly clusterProfiler package 17 (for enrichment analysis); org.Hs.eg.db package (version 3.10.0) (for ID conversion). Species: Homo sapiens.
Visualization
Software: R (version 3.6.3). R packages: mainly the ggplot2 package (for visualisation) || clusterProfiler package.
Statistical Analysis
Statistical analysis was performed according to the reference.18
Results
Clinical Characteristics
As shown in Table 1, there were 496 patients in this study. The FIGO stage included 1 T1 (0.2%), 29 T2 (5.9%), 382 T3 (77.6%), and 80 T4 (16.3%). The primary therapy outcome included 36 PD (8.8%), 25 SD (6.1%), 56 PR (13.7%), and 291 CR (71.3%). The race included 16 Asian (3.3%), 32 Black or African American (6.7%), and 431 White (90%). The age included 274 patients (≤60, 55.2%), and 222 patients (>60, 44.8%). The histological grade included 1 G1 (0.2%), 61 G2 (12.6%), 420 G3 (87%), and 1 G4 (0.2%). The anatomic neoplasm subdivision included 202 unilateral (26.6%) and 343 right (73.4%). The venous invasion included 75 yes (56.8%) and 57 no (43.2%). The lymphatic invasion included 116 yes (63.4%) and 67 no (36.6%). The tumor residual included 96 NRD (22%) and 341 RD (78%). The age range was 51 to 69 years, with a median of 59 years.
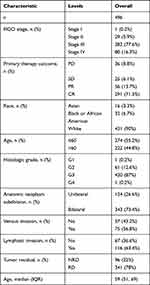 |
Table 1 Characteristics of Patients with OC Based on TCGA |
miR-501-3p Expression is Correlated with Poor Clinical Characteristics of OC
As shown in Figure 1, miR-501-3p expression was significantly lower in OC tissues (1.435 ± 0.093, n=10) compared with normal ovarian tissues (0.370 ± 0.033, n=10) (P<0.001). As shown in Table 2, miR-501-3p expression was associated with histological grade (P=0.015). As shown in Figure 2 and Table 3, miR-501-3p expression was significantly related to histological grade (P=0.015) and venous invasion (P=0.049). Collectively, the results suggested that high miR-501-3p expression was associated with the unfavorable prognosis among patients with OC.
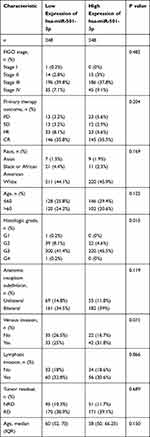 |
Table 2 Relationship Between miR-501-3p Expression and Clinical Characteristics in OC |
 |
Table 3 MiR-501-3p Expression Associated with Clinical Characteristics (Logistic Regression) |
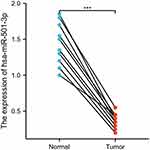 |
Figure 1 Expression of miR-501-3p in OC and normal tissues. Significance marker: *** p<0.001. |
 |
Figure 2 Association with miR-501-3p and clinical characteristics in OC. (A) Histological grade, (B) Venous invasion. Significance markers: *p< 0.05; **p<0.01. |
Role of miR-501-3p in OC Patient Survival
As shown in Figure 3, the expression of miR-501-3p was correlated with poor OS (HR: 0.77; 95% CI: 0.61–0.96; P=0.02) and DSS (HR: 0.77; 95% CI: 0.61–0.99; P=0.038). The above data indicated that miR-501-3p is a prognostic factor and low miR-501-3p level is associated with poor OS.
 |
Figure 3 Relationship between miR-501-3p expression and prognosis. (A) Overall survival, OS, (B) Disease specific survival, DSS. |
GO and KEGG Analysis of miR-501-3p in OC Patient Survival
The 793 target genes of miR-501-3p were listed in Table S3. The 986 upregulated genes of OC were listed in Table S4. The 35 common genes included AHRR, ALDH1L2, AURKB, BSPRY, COL12A1, COL23A1, COL5A1, DENND4B, DPEP3, E2F1, E2F7, ECT2, EZH2, FJX1, GAN, GPR160, HNRNPU, KCNT1, KLHL14, MAPKAPK2, MCM4, MTHFD2, MYO3B, OIP5, OLIG2, ORC6, RC3H1, SCRIB, SKIL, SLC39A14, SLC6A8, SSTR1, STK38, SYK, and TIMM8A (Figure 4). As shown in Table S5 and Figure 5, the common genes are involved in the biological process, including negative regulation of cell division, positive regulation of cell cycle process, attachment of spindle microtubules to kinetochore, positive regulation of cell cycle, and mRNA stabilization; cellular components including mitotic spindle midzone, chromocenter, chromosomal region, collagen trimer, and mitotic spindle; molecular function including extracellular matrix structural constituent conferring tensile strength, oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor, DNA replication origin binding, protein serine/threonine kinase activity, and oxidoreductase activity, acting on the CH-NH group of donors. As shown in Table S5 and Figure 6, the common genes are involved in the pathways including one carbon pool by folate, protein digestion and absorption, cell cycle, kaposi sarcoma-associated herpesvirus infection, and viral carcinogenesis.
 |
Figure 4 Venn diagram of common genes between target genes of miR-501-3p and LUAD down-regulated genes. |
 |
Figure 5 GO analysis of miR-501-3p target genes. Abbreviations: BP, biological process; CC, cellular component; MF, Molecular Function. |
 |
Figure 6 KEGG analysis of miR-501-3p target genes. |
Discussion
Aberrant expression of miRNAs is associated with diagnosis or prognosis in OC. MiRNA-150 can be a prognostic factor in advanced OC.19 The combined monitoring of serum miR-26b and miR-21 is of value in the clinical management of OC.20 MiR-27a is an oncogenic molecule and prognostic factor in patients with OC.21 Inhibition of miR-27a expression may be a new strategy for the treatment of OC.22 Therefore, the development of novel biomarkers for OC is of great importance.
MiR-501-3p was highly expressed in colorectal cancer tissues compared to normal colorectal tissues.23 MiR-501-3p expression was significantly down-regulated in prostate cancer tissues and cell lines.24 MiR-501-3p was lowly expressed in NSCLC tissues and cell lines compared to normal lung tissue.9 miR-501-3p was lowly expressed in HCC tissue samples, and downregulation of miR-501-3p was associated with tumor progression and poor prognosis in HCC patients.10 Expression of miR-501-3p was significantly upregulated in OS tissues and cell lines, and upregulated miR-501-3p expression was associated with TNM staging, distal metastasis and poorer prognosis in OS patients.11 MiR-501-3p expression was significantly lower in OC tissues compared with normal ovarian tissues (P<0.001). MiR-501-3p expression was significantly related to histological grade (P=0.015) and venous invasion (P=0.049). OC expressed less miR-501-3p than adjacent normal ovarian tissue, especially in patients with histologic grade G1&G2 compared to G3&G4. These phenomena suggest that miR-501-3p plays a role in the development of OC. Expression of miR-501-3p was higher in OC patients with venous invasion compared to OC patients with no venous invasion. The reason for this phenomenon is subject to further study. Low miR-501-3p expression predicted a poorer OS (HR: 0.77; 95% CI: 0.61–0.96; P=0.02) and DSS (HR: 0.77; 95% CI: 0.61–0.99; P=0.038).
MiR-501-3p is a novel regulator of Wnt/β-catenin signaling in CRC cells.23 MiR-501-3p limits prostate cancer cell growth by targeting CREPT to inhibit cell cycle protein D1 expression.24 MiR-501-3p acts as a tumor suppressor in NSCLC by directly targeting RAP1A mRNA.9 The M2 macrophage-derived exosome miR-501-3p suppresses the oncogenic TGFBR3 gene and promotes PDAC through activation of the TGF-β signaling pathway.5 MiR-501-3p inhibits metastasis and progression of HCC by targeting LIN7A.10 MiR-501-3p targets BCL7A to promote proliferation, migration, and invasion of osteosarcoma cells.11 Circ-ZNF609 accelerates glycolysis through the miR-501-3p/HK2 axis to promote PCa cell progression and radioresistance.25 In this study, the target genes of miR-501-3p are involved in the pathways including one carbon pool by folate, protein digestion and absorption, cell cycle, kaposi sarcoma-associated herpesvirus infection, and viral carcinogenesis.
This study has limitations as the analysis was limited to data sourced from databases and bioinformatics analysis. The clinical significance and specific molecular mechanisms of miR-501-3p in patients with OC in the real world need further investigation.
Conclusion
MiR-501-3p showed low expression in OC tissues. Low expression of MiR-501-3p was associated with poor OS and DSS in OC patients. MiR-501-3p might participate in the development of OC by pathways including small cell lung cancer, measles, PI3K-Akt signaling pathway, glioma, and p53 signaling pathway. MiR-501-3p may be a promising prognostic biomarker for OC patients.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Acknowledgments
The datasets generated in this study are available from TCGA that provide free resources.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by Jiangsu Provincial Key Discipline of Maternal and Child Health (2017103033).
Disclosure
The authors declare that they have no conflicts of interest relevant to this study.
References
1. Marth C, Reimer D, Zeimet AG. Front-line therapy of advanced epithelial ovarian cancer: standard treatment. Ann Oncol. 2017;28(suppl_8):viii36–viii39. doi:10.1093/annonc/mdx450
2. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386.
3. Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10(4):211–224.
4. Yin Z, Ma T, Huang B, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. 2019;38(1):310.
5. Lou W, Ding B, Zhong G, Du C, Fan W, Fu P. Dysregulation of pseudogene/lncRNA-hsa-miR-363-3p-SPOCK2 pathway fuels stage progression of ovarian cancer. Aging. 2019;11(23):11416–11439.
6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
7. Ma LL, Liang L, Zhou D, Wang SW. Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma. 2021;68(1):165–173.
8. Lu X, Li G, Liu S, Wang H, Chen B. MiR-585-3p suppresses tumor proliferation and migration by directly targeting CAPN9 in high grade serous ovarian cancer. J Ovarian Res. 2021;14(1):90.
9. Lu J, Zhou L, Wu B, et al. MiR-501-3p functions as a tumor suppressor in non-small cell lung cancer by downregulating RAP1A. Exp Cell Res. 2020;387(1):111752.
10. Luo C, Yin D, Zhan H, et al. microRNA-501-3p suppresses metastasis and progression of hepatocellular carcinoma through targeting LIN7A. Cell Death Dis. 2018;9(5):535.
11. Dai J, Lu L, Kang L, Zhang J. MiR-501-3p promotes osteosarcoma cell proliferation, migration and invasion by targeting BCL7A. Hum Cell. 2021;34(2):624–633.
12. Yuan J, Li T, Yi K, Hou M. The suppressive role of miR-362-3p in epithelial ovarian cancer. Heliyon. 2020;6(7):e04258.
13. Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.
14. Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009.
15. Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18.
16. Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500.
17. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287.
18. Lu X, Li G, Liu S, Wang H, Zhang Z, Chen B. Bioinformatics analysis of KIF1A expression and gene regulation network in ovarian carcinoma. Int J Gen Med. 2021;14:3707–3717.
19. Wilczyński M, Żytko E, Danielska J, et al. Clinical significance of miRNA-21, −103, −129, −150 in serous ovarian cancer. Arch Gynecol Obstet. 2018;297(3):741–748.
20. Song KW, Zhang QG, Tan WB, Fang YN. Diagnostic significance of serum miR-26b and miR-21 expressions in ovarian cancer and their associations with clinicopathological characteristics and prognosis of patients. Eur Rev Med Pharmacol Sci. 2020;24(4):1697–1703.
21. Wang Z, Ji G, Wu Q, et al. Integrated microarray meta-analysis identifies miRNA-27a as an oncogene in ovarian cancer by inhibiting FOXO1. Life Sci. 2018;210:263–270.
22. Teng Y, Su X, Zhang X, et al. miRNA-200a/c as potential biomarker in epithelial ovarian cancer (EOC): evidence based on miRNA meta-signature and clinical investigations. Oncotarget. 2016;7(49):81621–81633.
23. Wu F, Xing T, Gao X, Liu F. miR‑501‑3p promotes colorectal cancer progression via activation of Wnt/β‑catenin signaling. Int J Oncol. 2019;55(3):671–683.
24. Zhang Z, Shao L, Wang Y, Luo X. MicroRNA-501-3p restricts prostate cancer growth through regulating cell cycle-related and expression-elevated protein in tumor/cyclin D1 signaling. Biochem Biophys Res Commun. 2019;509(3):746–752.
25. Du S, Zhang P, Ren W, Yang F, Du C. Circ-ZNF609 accelerates the radioresistance of prostate cancer cells by promoting the glycolytic metabolism through miR-501-3p/HK2 axis. Cancer Manag Res. 2020;12:7487–7499.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
