Back to Journals » International Journal of General Medicine » Volume 15
Expression and Significance of Cyclin-Dependent Protein Kinase 6 in Diffuse Large B-Cell Lymphoma
Authors Li J, Li P, Su H, Feng H, Bai Z, Xi Y
Received 1 July 2022
Accepted for publication 22 August 2022
Published 14 September 2022 Volume 2022:15 Pages 7265—7276
DOI https://doi.org/10.2147/IJGM.S380496
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jing Li,1 Peng Li,2 Hong Su,1 Haonan Feng,3 Zhongyuan Bai,4 Yanfeng Xi1
1Department of Pathology, Cancer Hospital Affiliated to Shanxi Medical University, Shanxi Province Cancer Hospital, Shanxi Hospital Affiliated to Cancer Hospital, Chinese Academy of Medical Sciences, Taiyuan, Shanxi, 030013, People’s Republic of China; 2Department of Breast Surgery, Cancer Hospital Affiliated to Shanxi Medical University, Shanxi Province Cancer Hospital, Shanxi Hospital Affiliated to Cancer Hospital, Chinese Academy of Medical Sciences, Taiyuan, Shanxi, 030013, People’s Republic of China; 3Department of Pathology, Chengdu Second People’s Hospital, Chengdu, Sichuan, 610000, People’s Republic of China; 4Department of Pathology, Shanxi Medical University, Taiyuan, Shanxi, 030001, People’s Republic of China
Correspondence: Yanfeng Xi, Department of Pathology, Cancer Hospital Affiliated to Shanxi Medical University, Shanxi Province Cancer Hospital, Shanxi Hospital Affiliated to Cancer Hospital, Chinese Academy of Medical Sciences, 3 Workers’ New Village Street, Xing Hua Ling, Taiyuan, Shanxi, 030013, People’s Republic of China, Email [email protected]
Objective: To study the relationship between cyclin-dependent protein kinase 6 (CDK6) expression in diffuse large B-cell lymphoma (DLBCL) and the clinical biological behavior and prognosis.
Methods: Data mining was performed using the Oncomine and The Cancer Genome Atlas (TCGA) databases to analyze the expression level of CDK6 in DLBCL. CDK6 alterations in DLBCL and related functional networks were analyzed with c-BioPortal and the Gene Set Enrichment Analysis was performed by using DAVID and FunRich software. In addition, screening for differential gene expression of CDK6 was done and enriched by using LinkedOmics. Finally, formalin-fixed and paraffin-embedded (FFPE) tissue samples from 102 patients with DLBCL were collected from the Department of Pathology, Shanxi Cancer Hospital (Taiyuan, Shanxi, China) from January 2015 through December 2020. All cases had complete clinical course records. Thirty cases of lymph node reactive hyperplasia tissues were used as controls. The expression of CDK6 in DLBCL tissues was detected by qRT‑PCR and immunohistochemistry.
Results: Bioinformatics analysis: The data showed that mRNA expression level and DNA copy number variations (CNVs) of CDK6 were significantly higher in DLBCL as compared to normal tissue (P ˂ 0.05). Based on C-BioPortal analysis, we speculated that amplification was the most common copy of CDK6 CNV in DLBCL. Through Gene Ontology (GO) analysis of these genes, it was found that the proteins were mainly located in the nucleus and cytoplasm. The biological interaction network of CDK6 alterations were found to participate primarily in the G1-S phase of the process. Analysis of LinkedOmics mRNA sequencing data showed that three genes were positively correlated with CDK6 expression: PSMD1, C2orf29 and ASB1. Through experimental verification, we found that CDK6 was overexpressed in DLBCL, and the expression of CDK6 mRNA and protein in DLBCL were positively correlated with Ann Arbor staging and IPI score (P< 0.05), and negatively correlated with overall survival (P< 0.001).
Conclusion: Data mining results and experiments revealed and confirmed multi-level evidence for the importance of CDK6 in DLBCL; hence, CDK6 may be a potential marker in DLBCL. Thus, our study will perhaps lay the foundation for further research on the role of CDK6 in the genesis and development of DLBCL.
Keywords: cyclin-dependent protein kinase 6, diffuse large B-cell lymphoma, bioinformatics analysis, prognosis
Introduction
Diffuse large B-cell lymphoma (DLBCL), a type of non-Hodgkin lymphoma (NHL) originating from mature B cells, is characterized by high heterogeneity and invasiveness. It is the most common subtype of NHL, accounting for about 35–40% of NHL.1 DLBCL can be primary (inside or outside the lymph nodes), and can also be secondary to other low-grade malignant lymphomas. The typical manifestation is a rapidly growing mass inside or outside the lymph nodes, which can be accompanied by fever and other symptoms. Currently, more than 60% of DLBCL patients have significantly improved survival after treatment with the standard R-CHOP regimen (Rituximab combined with Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone); however, 30% to 40% of patients have relapsed or refractory disease, with an overall poor prognosis.2 Therefore, it is pertinent to search for new therapeutic targets for patients with DLBCL, so as to prolong the survival time and improve the prognosis of patients. Recently, molecular/cytogenetic profiling studies have independently identified 5–7 new functional genetic subgroups of DLBCL, strongly emphasizing the validity of this concept, but failing to classify all cases. A combination of cell-of-origin and molecular subclassification may provide more precise patient stratification for developing future clinical trials. Overall, cell-of-origin is retained for the present time with the expectation that transition to a molecular genetic classification will be feasible in the near future.1
One of the main causes of human tumorigenesis is the abnormal regulation of the cell cycle,3 which can be greatly affected by cyclin-dependent protein kinase (CDKs). As one of the important members of the CDK family, CDK6 plays a key role in the cell cycle process. It has serine/threonine kinase activity and normally works with cyclins to phosphorylate key proteins in the cell cycle. It shares 70% amino acid homology with cyclin-dependent protein kinase 4 (CDK4) and they have similar biochemical characteristics. CDK6 and CDK4 usually work in tandem to facilitate the transition of the cell cycle from the G1 to S phase.4,5 Several reports indicate that CDK6 not only plays an important role in the development of cells,6 but also participates in the occurrence and development of various malignant tumors.7–13 All relevant studies indicate that CDK6 is closely related to tumorigenesis.
In recent years, the role of CDK6 in malignant hematologic tumors has become a focus of research. Therefore, to determine the expression of CDK6 in DLBCL, the expression and mutation levels of CDK6 were explored in DLBCL patients in public databases, such as Oncomine. Using multidimensional analysis, the genomic alterations and functional networks were analyzed and correlated with CDK6 in DLBCL. Then, by screening the gene network related to the formation and progress of DLBCL involved in CDK6, the correlation between CDK6 and the mechanism of DLBCL was investigated. Finally, the mRNA and protein expression levels of CDK6 were detected in DLBCL patients by qRT-PCR and immunohistochemistry, respectively, and we found a correlation between CDK6 expression and clinicopathological characteristics and prognosis of DLBCL patients. Our results will provide new strategies for the early diagnosis and precise treatment of DLBCL and promote research on the mechanism of occurrence and development of DLBCL. Meanwhile, Predictive modeling is a feasible analytic strategy that results in a solution that can be understood.14
Materials and Methods
Bioinformatics Analysis
Oncomine Analysis
Oncomine (www.oncomine.org) is the world’s largest oncogene microarray database, which contains 715 gene expression datasets from 86,733 cancerous and normal tissues, facilitating comprehensive data mining.15 By analyzing the expression level and DNA copy number of CDK6 mRNA in DLBCL in the Oncomine 4.5 database, we considered relevant data from a series of DLBCL studies, including the Rosenwald, Brune, Storz, and TCGA lymphoma studies.16–18 The expression level was evaluated with P < 0.05 taken as a statistically significant difference.
c-BioPortal Analysis
The cBio Cancer Genomics Portal (http://cbioportal.org), which currently contains 225 tumor studies, is a cancer genomics portal with open access to resources, visualization, and multidimensional analysis of tumor genomic data.19 It was used to analyze CDK6 mutation levels in DLBCL samples from TCGA database, and the search parameters included mutations, copy number variations (CNVs), and mRNA expression. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of related genes with a frequency greater than zero were performed using Fun-Rich software.
DAVID Analysis
DAVID (https://david-d.ncifcrf.gov/) is a large-scale database that provides systematic and comprehensive biological function annotation information for large-scale gene or protein lists, helping users extract biological information from it. Through the DAVID 6.7 database, we analyzed the KEGG functional annotation map of 38 related genes centered on CDK6.
LinkedOmics Analysis
LinkedOmics (http://www.linkes.org/login.php) is an online analysis platform for tumor-related data.20 The LinkFinder module of LinkedOmics was used to study the differentially expressed genes (n = 48) related to CDK6 in DLCBL in TCGA database, and further statistical analysis was performed.
Exploration Experiment
Patients
(1) Formalin-fixed and paraffin-embedded (FFPE) tissue samples were collected from 102 patients, all of whom were diagnosed as DLBCL in the Department of Pathology, Shanxi Cancer Hospital (Taiyuan, Shanxi, China) from January 2015 through December 2020. All cases had complete clinical course records. Case inclusion criteria included the following: All patients had primary disease, had no previous history of tumor, and had not received radiotherapy or chemotherapy before surgery. (2) In addition, 30 cases of lymph node reactive hyperplasia tissues were used as controls. The use of human FFPE tissues was approved by the Shanxi Cancer Hospital Ethics Committee (no. 201964), and patient consent was obtained.
qRT‑PCR
Total RNA was extracted from FFPE tissue samples using TRIzol reagent (Xiamen Aide Biomedical Technology Co., LTD, Xiamen, China) strictly following the manufacturer’s instructions. CDK6 mRNA expression was quantified using the SYBR TB Green Premix Ex Taq II (Tli RNsesH Plus) (Bao Biological Engineering Co., LTD, Dalian, China) and normalized to β-actin expression using the 2−ΔΔCt method.21 The PCR primers used were as follows: CDK6 forward, 5’-AGAAGAAGACTGGCCTAGAG-3’ and reverse, 5’-TGGAAGTATGGGTGAGACAGG-3’; β-actin forward, 5’-CACCCAGCACAATGAAGAT-3’ and reverse, 5’-CAAATAAAGCCATGCCAAT-3’. All qRT‑PCR amplifications were performed in triplicate.
Immunohistochemistry
CDK6 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) immunohistochemical staining was performed on the tissue microarray. The positive signals for CDK6 were localized to the nucleus and cytoplasm; expression in either nucleus or cytoplasm in the sample was taken as positive (+). Results were double-blindly interpreted by at least two pathologists at the attending level or above. The CDK6 staining results were independently evaluated according to the interpretation standard of SU.22 The staining results were scored in terms of staining intensity of positive tumor cells and percentage of all tumor cells: No staining (score = 0), weak staining (score = 1), moderate staining (score = 2), strong staining (score = 3); we determined the mean percentage of positive tumor cells in at least five regions at ×40 magnification: 0% (score = 0), 1–30% (score = 1), 31–60% (score = 2), and > 60% (score = 3). The product of the two scores is the final interpretation score, with 6 as the critical value; < 6 as negative, and > 6 as positive. CD3, CD5, CD10, CD20, BCL-6, BCL-2, MUM1 and EBER were from Abcam. They were working fluid. The positive signals of CD3, CD5, CD10, CD20 was localized to cytoplasm. The positive signals of BCL-6, BCL-2, MUM1 and EBER was localized to the nucleus.
Statistical Analysis
SPSS25.0 software was used for statistical analysis. Statistical data analysis was performed using the Mann–Whitney U-test and the x2. Spearman rank correlation analysis was performed. Survival prognostic analysis was performed by the Kaplan–Meier method and GraphPad Prism 6.01 was used for graphing; P < 0.05 was considered statistically significant.
Results
Bioinformatics Analysis: CDK6 is Overexpressed in DLBCL and is Involved in Cell Cycle Regulation
Expression of CDK6 in DLBCL
We identified DLBCL studies using the Oncomine and TCGA databases and evaluated the CDK6 transcription level data contained therein. The data showed that the mRNA expression levels and DNA CNVs of CDK6 were significantly higher in DLBCL tissue compared to normal tissue (P ˂ 0.05). Although the fold change was within three, CDK6 was in the top 29% in gene sequencing based on mRNA expression, and in the top 10% in gene sequencing based on DNA CNVs (Figure 1).
Frequency and Type of CDK6 Alterations in DLBCL Patients
Next, we used c-BioPortal to analyze the frequency and type of CDK6 alterations based on the sequencing data of DLBCL patients from TCGA. CDK6 was altered in seven of the 48 (14.6%) patients with DLBCL (Figure 2A). The major alteration was amplification in two cases (4%). Therefore, we speculate that amplification is the most common CNV of CDK6 in DLBCL.
Biological Interaction Network of CDK6 Alterations in DLBCL
Next, we used network modules in c-BioPortal to identify 38 CDK6-related genes with frequency alterations (Figure 2B). The most frequently altered CDK6-related genes were CDKN2B (31.3%), CHEK1 (18.8%), and TP53 (16.7%). Through GO analysis of these genes, we found that the proteins they encoded were mainly located in the nucleus and cytoplasm. In addition, these proteins share serine/threonine protein kinase activity and participate in the regulation of the cell cycle (Figure 3A–C). KEGG pathway analysis showed enrichment mainly in the G1 phase and cyclin D-associated events in the G1 and mitotic G1-G1/S phases (Figure 3D and E). Therefore, CDK6 alterations mainly occur in the G1-S phase of the cell cycle.
Bioinformatics Analysis: CDK6 Overexpression in DLBCL is Specifically Associated with PSMD1, C2orf29, and ASB1
Using LinkedOmics, we analyzed the mRNA sequencing data of 48 DLBCL patients from TCGA. As can be seen in the volcano plot and related statistics, 566 genes (dark red spots) were significantly and positively correlated with CDK6, and 614 genes (dark green spots) were significantly and negatively correlated with CDK6 (Figure 4A and B). The top 50 genes positively and negatively associated with CDK6 are shown in the heat map (Figure 4C and D). The three most significant genes positively correlated with CDK6 expression were PSMD1 (Pearson correlation = 0.60, P = 8.18E-06), C2orf29 (Pearson correlation = 0.59, P = 8.39E-06), and ASB1 (Pearson correlation = 0.58, P = 1.78E-05) (Figure 5A–C).
Clinical Data
Of the 102 DLBCL patients, 58 were male and 44 were female. 78 cases were >60 years old, with an age range of 9 through 81 years, and a median age of 54 years. 57 cases had normal serum lactate dehydrogenase (LDH) levels (≤215 IU/L) and 45 cases had higher than normal (>215 IU/L) levels. The International Prognostic Index (IPI) scores were as follows: 76 cases had scores between 0–2 points and 26 cases between 3–5 points; the Ann Arbor staging was as follows: there were 51 cases in stage I–II, and 51 cases in stage III–IV. ECOG scores were 82 cases 1 point and 20 cases 2 point. There were 17 cases B symptom. 32 cases occurred extranodal sites (>1). According to Hans’ classifier, 40 cases were GCB and 62 cases were non-GCB (P>0.05). As of the end of June 2021, we followed up 102 DLBCL patients, of whom 63 survived and 39 died.
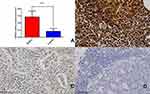
Immunohistochemistry Experimental Verification: CDK6 Expression Was Significantly Higher in DLBCL Tissues Compared with Reactive Lymph Node Hyperplasia Tissues
The ratio of CDK6 mRNA relative expression in 102 cases of DLBCL and 30 lymph node reactive hyperplasia controls was 0.546 (0.484, 0.649): 0.167 (0.107, 0.233). These results show that CDK6 mRNA was highly expressed in DLBCL tissues as compared to lymph node reactive hyperplasia, and the difference was statistically significant (P < 0.05) (Figure 6A). Immunohistochemical staining showed that, among 102 DLBCL tissue samples, the positivity rate of CDK6 was 55.88% (57/102)(including high expression and low expression) (Figure 6B and C). However, the positivity rate of CDK6 was 0 (0/30) in 30 samples of reactive hyperplasia of lymph nodes (Figure 6D). The differences were statistically significant (x2 = 29.506, P < 0.001) (Table 1). The positivity rate of CD3, CD5, CD10, CD20, BCL-6, BCL-2, MUM1 and EBER were 0.98%(1/102), 2.94%(3/102), 28.43%(29/102), 100%(102/102), 56.86%(58/102), 36.27%(37/102), 34.31%(35/102) and 4.90%(5/102)(P>0.05).
 |
Table 1 CDK6 Protein Expression in DLBCL and Reactive Hyperplasia of Lymph Nodes |
Correlation Between CDK6 Expression and Clinicopathological Features and Prognosis in DLBCL Patients
Statistical analysis showed that the CDK6 mRNA relative expression level was not significantly correlated with clinicopathological characteristics of DLBCL patients (P > 0.05), while the CDK6 protein expression was correlated with International Prognostic Index (IPI) and Ann Arbor staging (Table 2). Spearman rank correlation analysis showed that there was no significant correlation between the expression of CDK6 mRNA and protein in DLBCL (P > 0.05). The results of survival analysis showed that higher the level of CDK6 protein expression, the shorter was the overall survival of DLBCL patients (Figure 7, P < 0.001).
 |
Table 2 Relation of CDK6 Protein Expression with Clinical Feature of Patients with DLBCL |
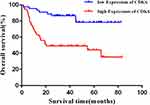 |
Figure 7 Upregulation of CDK6 protein expression predicts poor prognosis in DLBCL patients. |
Discussion
DLBCL refers to a type of NHL that originates from mature B cells and has high heterogeneity and invasiveness. Currently, there is a paucity of large-scale clinical research studies and corresponding effective treatment options. With the rapid development of tumor molecular biology technology, regulation of the tumor cell cycle at the molecular level and inhibition of tumor cell proliferation and growth have become hot subjects in oncology research in recent years. Various molecules, such as cyclin, CDK, CDK inhibitors (CDKI), and cell cycle checkpoints can cause the development of tumors through regulation of the cell cycle. CDK6, as a key cell cycle regulator, can also phosphorylate serine 536 of the p65 subunit of the nuclear transcription factor NF-κB and bind to the target gene promoter of NF-κB,23 further regulating the expression of oncogenes. Through extensive literature review, we found that CDK6 is involved in the occurrence and development of various malignant tumors, such as lung cancer,7 osteosarcoma,8 head and neck squamous cell cancer,9 bladder cancer,10 breast cancer,11 stomach cancer, oral cancer,12 ovarian cancer.13 In recent years, the role of CDK6 in malignant hematologic tumors has also become a focus of research. Current reports have confirmed that the overexpression of CDK6 is closely related to the development of lymphoma.3 In malignant tumors of the lymphatic system, CDK6 not only plays an important role in the regulation of cell cycle, but also induces angiogenesis. The study by Nagel et al24 highlights the critical role of CDK factor dysregulation in the pathogenesis of T-cell lymphoma. The results of Gu et al25 suggest that CDK6 may be a molecular therapeutic target for T-cell acute lymphoblastic leukemia. However, there are currently few studies on the expression of CDK6 in DLBCL patients. In 2019, Su et al22 found that down-regulation of miRNA-320d resulted in the loss of inhibition of cell proliferation, which further caused the overexpression of CDK6 in DLBCL patients. Therefore, CDK6 may affect the occurrence and development of DLBCL, but further verification is required.
In recent years, progress has been made in high-throughput technologies, such as second-generation sequencing and protein chips, with vast molecular data made available to the public. Thus, we can more conveniently and efficiently study the molecular basis of tumor occurrence and development. Moreover, these data will help find new gene targets for intervention and tumor treatment. To explore the expression of CDK6 in DLBCL and further study the mechanism of CDK6ʹs involvement in the occurrence and development of DLBCL, open sequencing data in several databases were analyzed by us with bioinformatics tools. We also experimentally verified the expression of CDK6 in DLBCL, with the aim of providing new strategies and targets for diagnosing and treating DLBCL.
By analyzing transcript sequencing data of DLBCL patients in Oncomine and TCGA databases, we found that CDK6 mRNA expression levels and CNVs were significantly higher than normal tissues. At the same time, according to the CNVs, we found that, although the fold change was not large, CDK6 ranked in the top 10% of all up-regulated genes in DLBCL. CNVs play an important role in the genome. They can disrupt and change the content of genes, leading to phenotypic differences.26 c-BioPortal analysis showed that CDK6 copies increased in DLBCL, with amplification as the main type of CDK6 change. We contend that changes and dysfunctions of CDK6 expression in DLBCL might be caused by changes to the chromosomal structure. Nevertheless, networks of related genes near CDK6 show various degrees of amplification and homozygous deletion in DLBCL. The relevant functional networks were involved in pre-DNA synthesis (G1 phase), cyclin D-associated events in G1, and the mitotic G1-G1/S phases. Meanwhile, the CDK6-related protein network was also enriched in PI3K/AKT, a classical signaling pathway. This is consistent with the physiological function of CDK6.27 Through enrichment analysis of CDK6-related differentially expressed genes, we found that the molecular functions of these genes are associated with serine/threonine kinase activity and are mainly involved in the regulation of cell cycle, which is also consistent with the mutation network.
Based on the above multidimensional bioinformatics analysis of CDK6 expression in DLBCL patients, we detected CDK6 expression in 102 DLBCL tissues at molecular and protein levels. The results showed that CDK6 was overexpressed at both molecular and protein levels in DLBCL patients, which was consistent with the results of bioinformatics analysis. Further, we analyzed the correlation between the expression levels of CDK6 and the clinicopathological characteristics and prognoses of DLBCL patients. The overexpression of CDK6 protein was not correlated with age, gender, LDH levels, etc., in patients with DLBCL. However, protein expression was positively correlated with the Ann Arbor staging and IPI of patients. The overexpression of CDK6 mRNA was not correlated with any of the patients’ clinical characteristics. DLBCL patients with positive CDK6 protein expression had shorter survival and poorer prognoses. The above results suggest that the abnormal expression of CDK6 may play an important role in the occurrence and development of DLBCL and portends a poor prognosis in these patients. Surprisingly, when combined with the immunohistochemical results, we found that the relative expression of CDK6 mRNA was not consistent with its protein expression. We speculate that the reasons for this discrepancy may be as follows:1) The selection of samples in qRT-PCR experiments has limitations; 2) Gene expression regulation is a complex process with multiple levels, stages, and modes. The process of gene expression is subject to stringent regulation, and the regulation of any one link will cause changes in the level of gene expression products; 3) At the same time, factors such as poor mRNA stability, faster degradation rate, and folding modification will also lead to inconsistencies between mRNA expression levels and protein expression levels.28
Conclusion
Our study used popular online tools based on bioinformatics to analyze target genes from cancer data available in public databases. Further, we explored the expression and correlation of target genes through experiments, with the aim of providing new therapeutic targets for DLBCL. Finally, we found multi-level evidence for the importance of CDK6 in DLBCL and its potential role as a marker for DLBCL. At the same time, our results suggest that CDK6 overexpression in DLBCL has a profound effect on the cell cycle. However, further studies on the related mechanisms of CDK6 in DLBCL are needed to confirm and validate our results.
Abbreviations
DLBCL, diffuse large B-cell lymphoma; NHL, non-Hodgkin lymphoma; CDK6, cyclin-dependent protein kinase 6; CDK4, cyclin-dependent protein kinase 4; TCGA, the Cancer Genome Atlas; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LDH, layered double hydroxide; IPI, international prognostic index.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Statement of Ethics
All participants gave the informed consent before their inclusion in the study. The study protocols were conducted according to the principles of the Declaration of Helsinki and were approved by the Scientific and Medical Ethical Committee of the Shanxi Provincial Cancer Hospital.
Ethics Approval and Consent to Participate
The use of human tissues was approved by the Shanxi Provincial Cancer Hospital Ethics Committee (no. 201964), and patient consent was obtained.
Patient Consent for Publication
Publication of the clinical datasets in this study does not compromise anonymity, or confidentiality, or breach local data protection laws.
Acknowledgments
We are grateful to Peng Bu and Ning Gao for their technical assistance.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The present study was supported by the Science and Technology Department of Shanxi Province of China (grant no. 2018067). The present study was supported by Natural Science Foundation of Shanxi Province, P. R. China (No. 20210302124576).
Disclosure
The authors declare that there is no conflict of interest.
References
1. Campo E, Jaffe ES, Cook JR, et al.The international consensus classification of mature lymphoid neoplasms: a report from the Clinical Advisory Committee. Blood. 2022. doi:10.1182/blood.2022015851
2. Feugier P, Van Hoof A, Sebban C, et al. Long-term results of the R-CHOP study in the treatment of elderly patients with diffuse large B-cell lymphoma: a study by the Groupe d’Etude des Lymphomes de l’Adulte. J Clin Oncol. 2005;23:4117–4126. doi:10.1200/JCO.2005.09.131
3. Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–181. doi:10.1016/j.ccr.2013.07.012
4. Thomas JW, Lee-Lin SQ, Green ED. Human-mouse comparative mapping of the genomic region containing CDK6: localization of an evolutionary breakpoint. Mamm Genome. 1999;10(7):764–767. doi:10.1007/s003359901088
5. Schulze-Gahmen U, Kim SH. Structural basis for CDK6 activation by a virus-encoded cyclin. Nat Struct Biol. 2002;9:177–181. doi:10.1038/nsb756
6. Malumbres M, Sotillo R, Santamaria D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi:10.1016/j.cell.2004.08.002
7. Igarashi K, Masaki T, Shiratori Y, et al. Activation of cyclin D1-related kinase in human lung adenocarcinoma. Br J Cancer. 1999;81:705–711. doi:10.1038/sj.bjc.6690752
8. Zhu K, Liu L, Zhang J, et al. MiR-29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6. Protein Cell. 2016;7:434–444. doi:10.1007/s13238-016-0277-2
9. Poomsawat S, Sanguansin S, Punyasingh J, et al. Expression of cdk6 in head and neck squamous cell carcinoma. Clin Oral Investig. 2016;20:57–63. doi:10.1007/s00784-015-1482-8
10. Wang G, Zheng L, Yu Z, et al. Increased cyclin-dependent kinase 6 expression in bladder cancer. Oncol Lett. 2012;4:43–46. doi:10.3892/ol.2012.695
11. Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi:10.1016/j.tibs.2005.09.005
12. Feng L, Xie Y, Zhang H, et al. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi:10.1007/s12032-011-9823-1
13. Xia B, Yang S, Liu T, et al. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi:10.1186/s12943-015-0322-4
14. Carreras J, Kikuti YY, Roncador G, et al. High expression of caspase-8 associated with improved survival in diffuse large B-cell lymphoma: machine learning and artificial neural networks analyses. BioMedInformatics. 2021;1(1):18–46. doi:10.3390/biomedinformatics1010003
15. Shanker K-S, Vasudeva M, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112
16. Andreas R, George W, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi:10.1056/NEJMoa012914
17. Verena B, Enrico T, Ines P, et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205(10):2251–2268. doi:10.1084/jem.20080809
18. Matt R, van de Rijn M, Kim YH, et al. Gene expression profiles of cutaneous B cell lymphoma. J Invest Dermatol. 2003;120(5):865–870. doi:10.1046/j.1523-1747.2003.12142.x
19. Jianjiong G, Arman AB, Ugur D, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
20. Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx109020
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
22. Hong S, Jiang C, Mengwei X, et al. CDK6 overexpression resulted from microRNA‑320d downregulation promotes cell proliferation in diffuse large B‑cell lymphoma. Oncol Rep. 2019;42(1):321–327. doi:10.3892/or.2019.7144
23. Handschick K, Beuerlein K, Jurida L, et al. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-kappaB-dependent gene expression. Mol Cell. 2014;53:193–208. doi:10.1016/j.molcel.2013.12.002
24. Nagel S, Leich E, Quentmeier H, et al. Amplification at 7q22 targets cyclin-dependent kinase 6 in T-cell lymphoma. Leukemia. 2008;22(2):387–392. doi:10.1038/sj.leu.2405028
25. Gu J, Wang X, Zhang L, et al. Matrine suppresses cell growth of diffuse large B-cell lymphoma via inhibiting CaMKIIγ/c-Myc/CDK6 signaling pathway. BMC Complement Med Ther. 2021;21(1):163. doi:10.1186/s12906-021-03315-0
26. Eugene U, Hao C, Zilu Z, et al. Integrative pipeline for profiling DNA copy number and inferring tumor phylogeny. Bioinformatics. 2018;34(12):2126–2128. doi:10.1093/bioinformatics/bty057
27. Amit D, Nicolette S, Schlichting N, et al. CDK6 kinase activity is required for thymocyte development. Blood. 2011;117(23):6120–6131. doi:10.1182/blood-2010-08-300517
28. de Sousa Abreu R, Penalva LO, Marcotte EM, et al. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5(12):1512–1526. doi:10.1039/b908315d
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.