Back to Journals » OncoTargets and Therapy » Volume 13
Expression and Prognostic Value of Id-4 in Patients with Esophageal Squamous Cell Carcinoma
Authors Wang X, Lu Q, Fei X, Zhao Y, Shi B, Li C, Chen H
Received 11 September 2019
Accepted for publication 12 December 2019
Published 12 February 2020 Volume 2020:13 Pages 1225—1234
DOI https://doi.org/10.2147/OTT.S230678
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Xinyu Wang,* Qijue Lu,* Xiang Fei,* Yue Zhao, Bowen Shi, Chunguang Li, Hezhong Chen
Department of Thoracic Surgery, Changhai Hospital, Second Military Medical University, Shanghai 200433, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hezhong Chen; Chunguang Li
Department of Thoracic Surgery, Changhai Hospital, Second Military Medical University, Shanghai 200433, People’s Republic of China
Tel +86 186 2127 9206
Fax +86 21 5578 6638
Email [email protected]; [email protected]
Background: Our previous study demonstrated that Id-1 may promote the tumorigenicity of esophageal squamous cell carcinoma (ESCC). Id-4 is another member of Id family, which is rare to be studied in ESCC. In this study, we investigated the expression of Id-4 in human ESCC specimens and determined whether Id-4 expression was associated with the clinicopathologic characteristic and the prognosis of ESCC patients.
Methods: We examined Id-4 expression using immunohistochemistry in 92 ESCC tissues and adjacent normal tissues. The association between Id-4 expression and clinical parameters and survival was evaluated by statistical analysis. Cox regression analyses were conducted to identify prognostic factors associated with overall survival (OS). In addition, we explored the functional mechanism of Id-4 in ESCC.
Results: Id-4 expression was significantly downregulated in ESCC tissues compared with adjacent normal tissues. The expression of Id-4 was associated negatively with pT stage (p=0.002), AJCC stage (p=0.008) and histologic differentiation (p< 0.001). OS was more unfavorable in patients with low expression of Id-4 than those with high expression of ESCC patients (p=0.007). In subgroup analysis, low expression of Id-4 could reveal unfavorable OS of patients with pT1b/T2 stage (p=0.024) or with pN0/N1 stage (p=0.004). By univariate analysis, pT stage and Id-4 expression showed statistically significant associations with OS (p=0.025, p=0.01, respectively). By multivariate analysis, Id-4 expression was an independent prognostic factor in ESCC (p =0.038). In addition, we observed that Id-4 could decrease the levels of the p-Smad2, p-Smad3 and TGF-β 1 in both Eca109 and TE1 cells, indicating Id-4 may inactivate the TGF-β signaling pathway.
Conclusion: Low expression of Id-4 suggested unfavorable prognosis for ESCC patients and could identify the prognosis in patients of early-stage tumors. The potential mechanism for Id-4’s tumor suppressor role in ESCC may be related to its inhibitory effect on TGF-β signaling pathway. Thus, we believe that Id-4 may be a promising prognostic marker and a therapeutic target in ESCC.
Keywords: Id-4, esophageal squamous cell carcinoma, prognosis, TGF-β signaling pathway
Introduction
Esophageal cancer (EC) is the sixth most lethal malignant tumor and the eighth most common malignant tumor in the world.1 According to the data from the CONCORD-3, the five-year survival rate of EC among most countries is lower than 30%, except Japan and South korea.2 While esophageal adenocarcinoma (EAC) is the main histologic subtype in west countries, esophageal squamous cell carcinoma (ESCC) remains the predominant form in China, accounting for more than 90% of EC.3 Surgery combined with chemoradiotherapy is the main treatment for ESCC. Despite advances in diagnosis and therapy, the five-year survival rate of patients with ESCC remains poor, which is mainly associated with recurrence and metastasis.4 Therefore, it is imperative to explore some molecular biomarkers associated with tumor tumorigenesis and progression of ESCC, aiming to predict tumor carcinogenesis and assess the prognosis for ESCC patients.
 |
Figure 3 Immunochemistry analysis of Id-4 expression. Representative staining intensity of Id-4 represented different expression levels in ESCC tissues and adjacent normal tissues. |
 |
Figure 4 The prognostic value of Id-4 in ESCC. Overall survival (OS) curves of Id-4 expression in patients with (A) primary tumors, with (B) pT1b-T2 stage, and with (C) N0-N1 stage. |
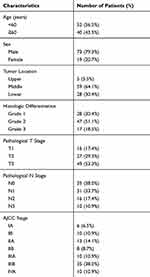 |
Table 1 Clinicopathologic Characteristics of 92 ESCC Patients |
 |
Table 2 Expression of Id-4 in ESCC Tumor Tissues and Corresponding Adjacent Normal Tissues |
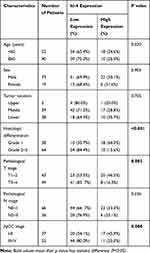 |
Table 3 Correlations Between Id-4 Protein Expression and Clinicopathologic Features |
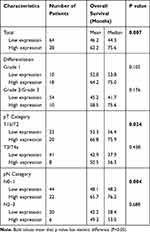 |
Table 4 Overall Survival Analysis According to Id-4 Expression in Patients with ESCC |
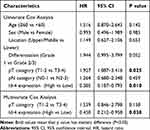 |
Table 5 Univariate and Multivariate Survival Analysis for Patients with ESCC |
The basic helix-loop-helix (bHLH) transcription factors are a large family of proteins that share a common HLH domain, which can be divided in seven classes. The inhibitor of DNA binding (Id) proteins belong to class V bHLH protein, including Id-1, Id-2, Id-3, and Id-4 members. As key regulators of cell cycle and differentiation, the expression of Id proteins is increasingly found in various cancers and is associated with poor prognosis, metastasis, angiogenesis, and epithelial-mesenchymal transition (EMT) in most cases.5
Our previous study demonstrated that Id-1 expression was positively correlated to LEF1 in ESCC patients, indicating Id-1 may promote the tumorigenicity of ESCC.6 Overall, the expressions and the underlying molecular mechanisms of Id-1, Id-2 and Id-3 in cancer have been studied thoroughly. However, Id-4 is rare to be studied in tumors, especially in ESCC. The biological function of Id-4 is different from other Id members, which has been considered as a tumor suppressor according to the existing studies.
Thus, we tried to investigate the Id-4 expression of human ESCC specimens. Meanwhile, we determined whether Id-4 expression was associated with the clinicopathologic characteristics and prognosis of ESCC patients.
Patients and Methods
Ethics Statement
The Ethics Committee of the Shanghai Changhai Hospital approved this study. All enrolled patients gave written informed consent before operation. All specimens were handled according to ethical and legal standards.
Patients Selection
Ninety-two consecutive patients with ESCC who had undergone esophagectomy from September 2012 to May 2013 at Shanghai Changhai Hospital were enrolled in this study. All patients included in the analysis fit the following criteria: (1) definite ESCC pathological results; (2) patients with complete resection of tumors (R0 resection); (3) patients were physically fit for surgery (normal functions of heart, lung, liver, kidney); (4) detailed and complete follow-up data. Exclusion criteria were conducted if any one of the following conditions was met: (1) history of other malignancies; (2) ESCC with T4b, M1 or recurrence; (3) preoperative radiotherapy or chemotherapy;
Follow-Up
Overall survival (OS) was defined as the interval between surgery and death or last observation. The follow-up was to conduct for 3-month intervals in the first year, 3- to 6-month intervals in the second year, and then once a year thereafter. After 5 years, consultation was implemented if the patients had any uncomfortable symptoms. All patients were followed up to June 2019.
Immunohistochemistry (IHC)
Paraffin-embedded tissues including normal and diseased regions from the 92 patients were consecutively resected into 5-µm thickness slices. These samples were deparaffinized and rehydrated. Epitope retrieval was performed with a heated citrate buffer for 30 mins. These sections were then incubated with primary mouse anti-human antibody against Id-4 (Abcam, ab49261, 1:100) overnight at 4°C. A biotinylated anti-rabbit secondary antibody was added for 30 min. Id-4 immunoreactivity was calculated by evaluating the percentage of positively-staining cells.
Bioinformatics Analysis
Oncomine database (https://www.oncomine.org/resource/login.html) and Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) were used for detecting the expression level of Id-4.
Cell Culture
Human esophageal epithelial cell (HEEC), Eca109, and TE1 cells were purchased from the Shanghai Cell Bank (Shanghai, China). Cells were cultured in DMEM (Gibco, CA, USA) supplemented with 10% fetal bovine serum (Gibco-BRL) and antibiotics (100 U/mL penicillin and 100 U/mL streptomycin, HyClone Laboratories, Inc., USA).
Western Blotting
In this method, 200 μL RIPA and 2.5 μL PMSF were mixed to collect protein for every 2.5*10^6 cells. Protein was separated by SDS-PAGE and transferred to PVDF membrane. Then, the membrane was incubated with the following primary antibodies: Id-4 antibody (Abcam, ab49261), TGF-β1 (Proteintech,21898-1-AP), p-Smad2(CST, 3108), p-Smad3(CST, 9520), Smad2(CST,5339), Smad3(CST, 9523) at 4°C for overnight. GAPDH antibody (Abcam, ab8245, 1:500) was used as a loading control. All experiments were performed in triplicates.
RNA Extraction and Quantitative RT-PCR (qRT-PCR)
The procedure was followed as described previously.7 Briefly, TRIzol (Invitrogen, Carlsbad, CA) was used to extract total RNA, and PrimeScript RT Reagent Kit (TaKaRa Bio, Shiga, Japan) was used to synthesize the cDNA. At last, SYBR Premix EX Taq (TaKaRa Bio, Shiga, Japan) was used for quantitative real-time polymerase chain reaction. Primer sequences are as follows.
Id-4: 5′-ATGAAGGCGGTGAGCCCGGT-3′ (forward)
5ʹ-AATGCTGTCGCCCTGCTTGTT-3ʹ (reverse);
GAPDH: 5ʹ-TCAAGAAGGTGGTGAAGCAG-3ʹ (forward)
5ʹ- GAGGGGAGATTCAGTGTGGT-3′ (reverse).
Statistical Methods
Statistical analysis was performed using the SPSS 24.0. Categorical variables were presented as percentages and were analyzed using the Chi-squared or Fisher’s exact tests. Continuous variables were expressed as mean ± standard deviation (SD). Kaplan–Meier curve and Log rank test were used to compare the OS between groups. Prognostic factors were determined by univariate and multivariate Cox regression analyses, and hazard ratio (HR) with 95% confidence interval (CI) were calculated. A significant difference was defined as p value < 0.05.
Results
Id-4 Is Downregulated in EC from the Published Database
First, we put the Id-4 gene into the Su Esophagus 2 and the Hu Esophagus database in Oncomine for detecting the expression level in ESCC. Then, we put the Id-4 gene into the other database, GEPIA. As a result, both of them showed that Id-4 expression in the tumor samples was lower than that in the normal tissues (Figure 1A and B).
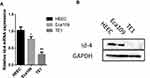
Comparison of Id-4 Expression Between ESCC Cells and HEEC
To further confirm the expression of Id-4, we compared the Id-4 expression level between ESCC cells (Eca109 cell and TE1 cell) and HEEC using qRT-PCR and Western blotting. Compared with that in the HEEC, our present study showed that Id-4 expression was lower in Eca109 and TE1 cells (Figure 2A and B).
Clinical and Pathologic Data
A total of 92 tumor samples with ESCC were collected in our study. There were 73 men and 19 women, with a mean age of 58.46±9.18 years. Follow-up data were collected from all patients, with a mean survival of 51.08 months (range, 5.5 to 80.2 months). Detailed clinical and pathologic data are shown in Table 1.
Associations Between Id-4 Expression and Clinicopathological Characteristics
Ninety-two pair of ESCC tissues and adjacent normal tissues were used for testing the levels of Id-4 by immunohistochemistry staining. The sections stained immunohistochemically for Id-4 protein were reviewed under a microscope and separately scored by two independent pathologists. The immunostaining staining levels were evaluated as 0 (negative), 1+ (weak positive), 2+ (moderate positive), and 3+ (strong positive). High expression level was defined as a score ≥2 (Figure 3).
As shown in Table 2, ESCC patients with high expression of Id-4 accounted for 30.4%. On the contrary, up to 65.2% of adjacent normal tissues showed high expression. Table 3 shows the correlation between Id-4 expression and clinicopathological features in ESCC patients. The results demonstrated that the expression of Id-4 was associated closely with pT stage (p=0.002) and AJCC stage (p=0.008). Especially, a significant association was observed between Id-4 expression and histologic differentiation (p<0.001). There was no association between Id-4 expression and age (p=0.320), sex (p=0.903), tumor location (p=0.705) or pN stage (p=0.336).
Correlation Between Id-4 Expression and Tumor Prognosis
By Kaplan–Meier analysis, overall survival (OS) was obviously more favorable in high expression of Id-4 than those with low expression (mean, 62.2 versus 46.2 months; median, 75.6 versus 44.3 months; p=0.007; Figure 4A).
By subgroup analysis (Table 4), the Kaplan–Meier analysis demonstrated that patients with low expression of Id-4 had unfavorable OS in pT1b/T2 stage (mean, 66.8 versus 52.3 months; median, 75.9 versus 56.4 months; p=0.024; Figure 4B) or with pN0/N1 stage (mean, 65.7 versus 48.1 months; median, 76.2 versus 48.2 months; p=0.004; Figure 4C).
As shown in Table 5, pT stage and Id-4 expression showed statistically significant associations with OS (p=0.025; p=0.01) by univariate Cox analysis. Next, we put both of them into multivariate analysis for detecting independent prognostic factors. By multivariate Cox analysis, only Id-4 expression showed a statistically significant association with OS (p =0.038).
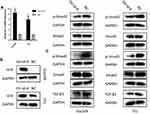
Id-4 May Suppress TGF-β Signaling Pathway in ESCC
To explore the molecular mechanism of Id-4 in ESCC, the overexpression plasmid of Id-4 was constructed by the Heyuan Biotechnology Company (Shanghai, China). The results of qRT-PCR and Western blotting showed that Id-4 protein level was increased significantly after transfection (Figure 5A and B). Besides, the key proteins of the TGF-β pathway were measured. The Western blotting results demonstrated that Id-4 could decrease the level of the p-Smad2, p-Smad3 and TGF-β1 in both ECA109 and TE1 cells while no changes in Smad2 and Smad3, indicating Id-4 may inhibit the TGF-β signaling pathway (Figure 5C).
Discussion
The expression of Id proteins is increasingly observed in various cancers. The Id proteins play a considerable role in the regulation of cell-cycle progression and cell differentiation by direct or indirect mechanisms.5
Although Id proteins are not classical oncogenes, they may regulate downstream target molecules and activate carcinogenic pathways. Id-1, Id-2 and Id-3, are universally believed as tumor supporting oncogenes.8 They contribute to tumorigenesis by inhibiting cell differentiation, promoting cell proliferation, facilitating tumour angiogenesis and promoting EMT.9 For example, high levels of Id-1 and the matrix metalloproteinases (MMPs) have been correlated with metastasis of breast cancer.10 The suppression of zinc-finger protein KLF17 results in Id1 activation, which promotes cell invasion and EMT of breast cancer.11 In ESCC, Li et al found that Id-1 could promote carcinogenesis and distant metastasis of ESCC through activating PI3K/AKT signaling pathway.12,13
However, Id-4 has been deemed as a tumor suppressor because it is epigenetically silenced in various cancers, such as cancer of prostate,14 breast,15 stomach,16 colorectum,17 and the hematopoietic and lymphoid tissues,18,19 as reported. Vitro studies have also demonstrated that ectopic Id-4 expression could inhibit proliferation, promote senescence, apoptosis and sensitivity to chemotherapeutic drugs.8 On the other hand, several studies demonstrated that Id-4 could also act as an oncogene in few cancers, such as brain tumors20 and ovarian cancer,21 which supports its role in promoting the tumorigenesis.22
The biological mechanism of Id-4 as a tumor suppressor is not understood fully. Aberrant promoter methylation of Id-4 followed by the decreased expression of Id-4 is one of the potential molecular mechanisms, which has been confirmed in prostate cancer,23 breast cancer,15 lung cancer,24 colorectal cancer,25 and lymphocytic leukemia.19 Besides, Pankaj Sharma et found that Id-4 could heterodimerize with Id-1, −2 and −3 with consequent reactivation of DNA transcription, thus antagonizing the biological activities of such Id proteins.8 In addition, Id-4 may collaborate with androgen receptor (AR) to exert the biological functions of carcinostasis.26
The expression of Id-4 in ESCC and the association of Id-4 with prognosis in ESCC patients have not been reported to our knowledge. In the present study, we firstly revealed that the low expression of Id-4 was found more frequently in patients with advanced pT stage (p=0.002), advanced AJCC stage (p=0.008) and poor differentiation (p<0.001). The poor prognosis of ESCC patients is mainly due to the advanced stage of cancer, poor histologic differentiation and high rates of regional lymph node metastasis.27 In this study, advanced pT stage and poor differentiation of ESCC patients with a low Id-4 expression may be responsible for the poor clinical outcome.
Compared with the advanced stage of tumor, early tumor is generally considered to have a better prognosis. However, many of these patients still suffer from tumor recurrence or metastasis, which causes a poor OS for such patients. Therefore, proper tumor biomarkers are imperatively needed to predict the prognosis for ESCC patients of early stage. By Kaplan–Meier analysis, we observed a significant difference in OS between the Id-4 low and high expression groups (p=0.024, p=0.004, respectively) among patients with early T and early N. Thus, the findings of the present study suggest that the measurement of Id-4 protein levels could identify the prognosis in patients with early-stage tumors.
TGF-β signaling pathway has been widely recognized for promoting the growth and metastasis of tumors by inducing EMT and tumor angiogenesis.28 Our previous study demonstrated that Id-1 and Id-3 were enriched in the TGF-β signaling pathway.6 Meanwhile, we found Id-4 may be involved in the TGF-β signaling pathway from Kyoto Encyclopedia of Genes and Genomes (KEEG) database. Thus, we believed Id family members could have a close link with the TGF-β signaling pathway in ESCC. In our study, we found that Id-4 could suppress the TGF-β signaling pathway, which might support the role of ID-4 as tumor suppressor.
In conclusion, Id-4 expression is significantly downregulated in ESCC tissues compared with adjacent normal tissues. ESCC patients with low Id-4 expression have worse OS. Our study suggests that Id-4 expression might be a prognostic biomarker and a potentially important therapeutic target of ESCC.
Consent for Publication
All the patients give consent for publishing their individual data.
Acknowledgment
We appreciated Xinyuan Lu and Rong Li from Eastern Hepatobiliary Surgery Hospital for reviewing the specimens and providing the suggestions of pathology.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81472688).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Allemani C, Matsuda T, Veronica Di C, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37513025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet. 2018;391(10125):1023–1075.
3. Domper Arnal MJ. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–7943. doi:10.3748/wjg.v21.i26.7933
4. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
5. Roschger C, Cabrele C. The Id-protein family in developmental and cancer-associated pathways. Cell Commun Signal. 2017;15(1):7.
6. Zhao Y, Zhu J, Shi B, et al. The transcription factor LEF1 promotes tumorigenicity and activates the TGF-β signaling pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2019;38(1):304. doi:10.1186/s13046-019-1296-7
7. Zhao Y, Li C, Huang L, et al. Prognostic value of association of OCT4 with LEF1 expression in esophageal squamous cell carcinoma and their impact on epithelial-mesenchymal transition, invasion, and migration. Cancer Med. 2018;7(8):3977–3987. doi:10.1002/cam4.2018.7.issue-8
8. Sharma P, Chinaranagari S, Chaudhary J. Inhibitor of differentiation 4 (Id-4) acts as an inhibitor of ID-1, −2 and −3 and promotes basic helix loop helix (bHLH) E47 DNA binding and transcriptional activity. Biochimie. 2015;112:139–150. doi:10.1016/j.biochi.2015.03.006
9. Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5(8):603–614. doi:10.1038/nrc1673
10. Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi:10.1038/nature03799
11. Gumireddy K, Li A, Gimotty PA, et al. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11(11):1297–1304. doi:10.1038/ncb1974
12. Li B, Cheung P, Wang X, et al. Id-1 activation of PI3K/Akt/NFκB signaling pathway and its significance in promoting survival of esophageal cancer cells. Carcinogenesis. 2007;28(11):2313–2320. doi:10.1093/carcin/bgm152
13. Li B, Tsao SW, Li YY, et al. Id-1 promotes tumorigenicity and metastasis of human esophageal cancer cells through activation of PI3K/AKT signaling pathway. Int J Cancer. 2010;125(11):2576–2585. doi:10.1002/ijc.24675
14. Sharma P, Chinaranagari S, Patel D, Carey J, Chaudhary J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Med. 2012;1(2):176–186. doi:10.1002/cam4.16
15. Nasif D, Campoy E, Laurito S, et al. Epigenetic regulation of ID4 in breast cancer: tumor suppressor or oncogene? Clin Epigenetics. 2018;10(1):111. doi:10.1186/s13148-018-0542-8
16. Chan AS, Tsui WY, Chen X, et al. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003;22(44):6946–6953. doi:10.1038/sj.onc.1206799
17. Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10(22):7475–7483. doi:10.1158/1078-0432.CCR-04-0689
18. Yu L, Liu C, Vandeusen J, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37(3):265–274. doi:10.1038/ng1521
19. Chen SS, Claus R, Lucas DM, et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117(3):862–871. doi:10.1182/blood-2010-05-284638
20. Zeng W, Rushing EJ, Hartmann DP, Azumi N. Increased inhibitor of differentiation 4 (id4) expression in glioblastoma: a tissue microarray study. J Cancer. 2010;1:1–5. doi:10.7150/jca.1.1
21. Ren Y, Cheung HW, von Maltzhan G, et al. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Sci Transl Med. 2012;4(147):147ra112. doi:10.1126/scitranslmed.3003778
22. Beger C, Pierce LN, Kruger M, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA. 2001;98(1):130–135. doi:10.1073/pnas.98.1.130
23. Vinarskaja A, Goering W, Ingenwerth M, Schulz WA. ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World J Urol. 2012;30(3):319–325. doi:10.1007/s00345-011-0750-8
24. Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi:10.1186/1479-5876-8-86
25. Borinstein SC, Conerly M, Dzieciatkowski S, et al. Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model. Mol Carcinog. 2010;49(1):94–103. doi:10.1002/mc.20581
26. Komaragiri SK, Bostanthirige DH, Morton DJ, et al. ID4 promotes AR expression and blocks tumorigenicity of PC3 prostate cancer cells. Biochem Biophys Res Commun. 2016;478(1):60–66. doi:10.1016/j.bbrc.2016.07.092
27. Yuequan J, Shifeng C, Bing Z. Prognostic factors and family history for survival of esophageal squamous cell carcinoma patients after surgery. Ann Thorac Surg. 2010;90(3):908–913. doi:10.1016/j.athoracsur.2010.05.060
28. Chen Y, Di C, Zhang X, et al. Transforming growth factor β signaling pathway: a promising therapeutic target for cancer. J Cell Physiol. 2019. Epub ahead of print.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.