Back to Journals » OncoTargets and Therapy » Volume 14
Expression and Functional Relevance of ANXA1 in Hypopharyngeal Carcinoma with Lymph Node Metastasis
Authors Li L, Wang Z, Lu T, Li Y, Pan M, Yu D, Hu G
Received 24 November 2020
Accepted for publication 11 February 2021
Published 25 February 2021 Volume 2021:14 Pages 1387—1399
DOI https://doi.org/10.2147/OTT.S292287
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Lei Li, Zhihai Wang, Tao Lu, Yanshi Li, Min Pan, Dan Yu, Guohua Hu
Department of Otolaryngology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Guohua Hu
Department of Otolaryngology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China
Tel +86-23-89012945
Fax +86-23-89012981
Email [email protected]
Purpose: The purpose of this study is to investigate the expression and functional role of Annexin (ANXA1) in lymph node (LN) metastasis of hypopharyngeal carcinoma (HSCC).
Methods: Differentially expressed genes in tissue from HSCC with or without LN metastasis were obtained from a previous RNA sequencing experiment. The presence of LN metastasis is determined by pathological diagnosis after neck dissection. ANXA1 expression was detected by qRT-PCR and Western blotting. Immunohistochemistry was used to detect the expression of ANXA1 in 74 cases of HSCC and normal control tissues. We also evaluated the clinical significance of ANXA1 in HSCC. Differentially expressed genes related to ANXA1 were analyzed using bioinformatic tools, and potential mechanisms of action of ANXA1 were assessed using in vitro experiments. In these in vitro experiments, cell proliferation was detected by CCK8 staining, and colony formation, migration and invasion were assessed using Transwell assays, and apoptosis as well as cell cycle status were quantified by flow cytometry.
Results: ANXA1 was significantly downregulated in HSCC with LN metastasis. The survival rate of patients with low ANXA1 expression was significantly worse than that of patients with high ANXA1 expression (p< 0.05). Silencing ANXA1 in cell culture experiments promoted the proliferation, migration and invasion of FaDu cells, inhibited apoptosis, and increased the proportion of cells in S phase. We furthermore found that the mRNA expression of ANXA1 was positively correlated with Yap1 expression (p< 0.0001). Our in vitro experiments showed that ANXA1 regulates the expression of Yap1, and over-expression of Yap1 could reverse the effect of ANXA1 silencing on cancer cell progression.
Conclusion: Our findings suggest that ANXA1 is a putative LN metastasis suppressor gene in tumor, which may suppress the LN metastasis of HSCC by regulating the expression of Yap1.
Keywords: HSCC, ANXA1, Yap1, LN metastasis
Introduction
Hypopharyngeal squamous cell carcinoma (HSCC) is a head and neck tumor with a high degree of malignancy and poses a serious threat to human life and health. Most patients with HSCC are diagnosed at advanced stages and 60–80% of HSCC patients have lymph node (LN) metastases at initial treatment.1–3 With advances in diagnosis and treatment, the primary tumor control of HSCC has reached a satisfactory level, yet the 5-year survival rate ranges only between 13–50%.4–6 Numerous studies have confirmed that HSCC with LN metastasis often results in treatment failure and represents an independent risk factor for survival.7–9 Therefore, it is important to study the potential molecular mechanisms affecting the malignant biological behavior of HSCC with LN metastasis in order to find more effective early diagnostic tools and reliable biomarkers.
Annexin (ANXA1) is the first characteristic member of the membrane-linked protein superfamily which was discovered to have anti-inflammatory properties. Numerous studies have since shown that ANXA1 is not only involved in many physiological processes, but also that aberrant expression of ANXA1 is closely related to tumorigenesis and tumor metastasis.10,11 ANXA1 has been shown to be up-regulated in breast,12 liver13 and colorectal cancer,14 while it is down-regulated in esophageal squamous cell carcinoma,15 prostate cancer16 and tumors of the oral cavity.17 Down-regulation of ANXA1 is strongly associated with a poor prognosis and distant metastasis of nasopharyngeal cancer.18 However, while many studies have reported an important role of ANXA1 in the development of tumors, its expression and potential roles in LN metastases of HSCC have not yet been investigated.
In this study, we found for the first time that ANXA1 plays a key role in LN metastasis of HSCC, and there is a significant correlation between low ANXA1 expression and poor prognosis of hypopharyngeal cancer patients. In addition, we preliminarily explored the mechanism of ANXA1 in HSCC and found that ANXA1 may suppress LN metastasis of HSCC by regulating Yap1.
Materials and Methods
Patients and Tissue Samples
The study was carried out in accordance with the Declaration of Helsinki and permission to carry out this study was obtained from the institutional review Ethics Committee of the first affiliated hospital of Chongqing Medical University. We screened patients diagnosed with HSCC in Otolaryngology of the First Affiliated Hospital of Chongqing Medical University between 2012 and 2019. The inclusion standards were as follows: (1) The pathological diagnosis was hypopharyngeal squamous cell carcinoma (HSCC). (2) Patients were treated in strict accordance with NCCN guidelines. (3) TNM was staged according to the AJCC 8th Edition. (3) All patients were examined by electronic nasopharyngoscope, CT or MRI every six months after the initial operation, and the follow-up records were complete. Exclusion criteria were as follows: (1) Patients with hypopharyngeal cancer who received preoperative radiotherapy or chemotherapy. (2) Patients with other tumors. Two experienced pathologists identified clinicopathological features of the specimens. Finally, 74 HSCC patients who met the inclusion criteria were selected. Each patient signed the relevant informed consent to protect their safety and privacy.
Cell Culture and Transfection
The FaDu cell line was purchased from cell bank of Chinese Academy of Sciences (Shanghai, China). Cells were grown in DMEM/high glucose medium (Hyclone, Logan, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Waltham, USA) and incubated in 5% CO2 at 37°C. si-NC and si-ANXA1 constructs were acquired from GenePharma (Shanghai, China). According to the manufacturer’s instructions, FaDu cells were inoculated into a 6-well plate at a density of 1 × 106 cells/well. When FaDu cells were in the logarithmic phase, the siRNA was transfected with liposomal RNA IMAX (Invitrogen, Carlsbad, CA, USA), and cells subsequently underwent incubation at 37°C. After 48 and 72 h, the mRNA and protein levels were detected by qRT-PCR and Western blotting.
Construction of Cell Line
In order to establish a stable ANXA1-silenced FaDu cell line, we purchased a lentivirus (sh-NC and sh-ANXA1) from GenePharma (Shanghai, China) according to the above sequence of the ANXA1 silenced group. As per the manufacturer’s instructions, the cells were seeded into six well plates at a density of 1 × 105 cells per well and cultured in a 37°C incubator for 16–24 h until the cells grew to 20–40% confluence. The multiplicity of infection (MOI) of the cells was 10. After 48–72 h of lentivirus infection (70–80% confluence), the cells were cultured in medium containing 2 μg/mL puromycin (Beyotime, China) until the control cells were killed by puromycin. Some cells were further screened and amplified, and then cells were collected for PCR and Western blotting identification. Similarly, we purchased viruses for Yap1 overexpression (oe-NC and oe-Yap1) from GENE (Shanghai, China) and transfected cells according to the same procedure as above.
Immunohistochemistry (IHC)
IHC was performed on 4 μm sections of paraffin-embedded tissues previously prepared. The prepared paraffin sections were dewaxed in fresh xylene and hydrated in gradient alcohol. Antigen repair was carried out with citrate buffer at 100°C for 30 min. Anti-ANXA1 (Cell Signaling Technology, Beverly, MA) was diluted 1:200 and the sections were incubated with the antibody at 4°C overnight. The next day, sections were washed three times on a shaker in PBS, Diaminobenzidine (DAB) reagents (ZSGB-BIO, China) were applied to detect the signal from the antigen-antibody reaction for 5 min. All sections were stained with hematoxylin for 60 s and washed with tap water for 10 min. Finally, the slides were dehydrated in gradient xylene and gradient alcohol, respectively, and were covered with a coverslip. Normal mucous tissue was selected as positive control. Phosphate buffer (PBS) was used to replace the primary antibody and incubated with sections as negative control. All sections were scored by two independent pathologists who were blinded to the clinicopathological information. Depending on the area and intensity of staining, the expression of ANXA1 was scored as either negative (no staining or only weak staining, ≤50% of tumor cells) or positive (moderate staining or strong staining, >50% of tumor cells).
Western Blotting (WB)
The total protein extraction kit (China KeyGen biotechnology company) was used to lyse hypopharyngeal cancer specimens and treated cells. The same amount of total protein was separated on a 10% SDS-PAGE, transferred to a PVDF membrane and blocked with 5% skim milk at room temperature for 1 h. Membranes were probed with the following primary antibodies overnight at 4°C: rabbit anti-human ANXA1 monoclonal antibody (1:1000, CST, Boston, Massachusetts USA); rabbit anti-human Yap1 polyclonal antibody (1:2000, Proteintech, Chicago, Illinois, USA); rabbit anti-human GAPDH monoclonal antibody (1:3000, Proteintech, Chicago, Illinois, USA) were used as internal reference antibodies. The secondary antibody goat anti-rabbit IgG (Beyotime, 1:5000, China) was added into the membrane and washed three times in PBST on a shaker for 10 min each time. The ECL system (Pierce, Thermo, USA) was utilized to detect immunoactivity.
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated from human tissues and cells using the Total RAN Kit I (Omega, USA) according to the manufacturer’s instructions. The primescript RT Kit (Takara, Dalian, China) was used for reverse transcription (RT) and SYBR primescript RT-PCR Kit (Takara, Dalian, China) was used for qRT-PCR. GAPDH served as an internal reference gene. The relative expression levels were calculated using the 2−ΔΔCt method. Primer sequences are listed in Table 1.
 |
Table 1 Primer Sequences |
Bioinformatics
In a previous study, we obtained the mRNA expression results of HSCC patients with LN metastasis and no LN metastasis by RNA sequencing. The edge software package was used to analyze the differentially expressed genes (DEmRNA) (|log2FC|>2, p<0.05) between the group of HSCC patients with or without LN metastasis group. The functional enrichment of ANXA1-correlated genes was analyzed by NetworkAnalyst (https://www.networkanalyst.ca/).19 Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified with a strict cutoff (false discovery rate (FDR)<0.05). The protein–protein interaction (PPI) for ANXA1 and correlated genes was analyzed by STRING (http://www.string-db.org/).20 The visual network of pivotal genes was constructed using the Cytoscape software.21
Cell Proliferation Assay
The cell proliferation reagent CCK-8 (Monmouth Junction, NJ, USA) was used to assess the proliferation of FaDu cells. In short, cells transfected as described above and incubated for 48 h were added to 96 well plates (1×103 cells/well) containing 10% FBS medium. Ten microliters (10 μL) of CCK-8 were added to cells and cells were incubated at 37°C for 1 h. Finally, the optical density at 450 nm was measured using a microplate reader (iMark Microplate Absorbance Reader; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Colony Formation Analysis
The colony-forming assay was used to detect cell proliferation. Forty-eight hours (48 h) after transfection, 1000 cells per space were inoculated into six empty plates and incubated in medium containing 10% FBS for about 2 weeks. The medium was changed every three days. Finally, cells were fixed with 100% formaldehyde and stained with 0.5% crystal violet. The number of colonies was counted under an inverted microscope.
Transwell Migration and Invasion Assay
Cell invasiveness and migration were measured a Transwell assay using 8 µm pore-chambers (Corning, USA). Briefly, a 2.5 × 105 cells/mL suspension was generated from treated cells using serum-free DMEM medium. Two hundred microliters (200 μL) cell suspension and 600 µL DMEM medium containing 10% FBS were added into the upper and lower chamber, respectively. After 24 h of incubation, the cells in the chamber were fixed and stained with 100% paraformaldehyde and 0.5% crystal violet. The cells in the upper chamber were removed and evaluated under a microscope. In order to further evaluate cell invasiveness, the assay was repeated in a Transwell assay with Matrigel (BD Biosciences, Bedford, MA, USA).
Assessment of Apoptosis
Apoptotic cells were labeled with the annexin V FITC/PI detection kit (SunGENE BioTEch, Tianjin, China). After 72 h of transfection, cells were trypsinized and washed with PBS. According to the manufacturer’s instructions, cells were labeled in a binding buffer containing annexin V-FITC/PI. The samples were analyzed by flow cytometry (FCM, BD Biosciences, USA).
Cell Cycle Analysis
After 48 h of transfection, the cells were digested with trypsin, washed twice with cold PBS, fixed with 70% ice ethanol, and overnight in a refrigerator at −4 °C. The cells were washed with cold PBS and treated with RNase A and propidium iodide according to the instructions. The cell cycle was measured by FCM.
Statistical Analysis
SPSS 22.0 and GraphPad Prism 6.0 were employed for statistical analysis. Group differences were assessed using the Student’s t-test for parametric test and the Mann–Whitney U-test for nonparametric test. The immunohistochemical expression difference of ANXA1 in patients with or without LN metastasis and the association of ANXA1 expression with HSCC clinicopathological features were evaluated using a Chi-square test. The Pearson test was used to analyze the correlation between ANXA1 and Yap1 expression. Kaplan–Meier and log-rank analysis were used to evaluate prognostic factors. All data are presented as mean ± standard deviation (SD), and a p-value<0.05 was considered statistically significant.
Results
ANXA1 Expression Correlates with LN Metastasis of HSCC
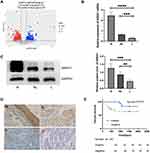
A total of 698 DEmRNAs were identified in the HSCC samples between patients with and without LN metastasis, including 278 upregulated genes and 420 downregulated genes (Figure 1A). ANXA1 was one of these DEmRNAs. To further confirm the expression of ANXA1 in this study, we increased the number of samples and also assessed ANXA1 protein levels. We found that mRNA levels of ANXA1 (Figure 1B) were significantly lower in the LN metastasis group than in the normal group (p<0.0001) and the HSCC patient group without LN metastasis (p<0.001). Protein expression levels of ANXA1 (Figure 1C) were significantly lower in the LN metastasis group than in the normal group (p<0.001) and the HSCC patient group without LN metastasis (p<0.01), suggesting that a lack of ANXA1 might be a key component of metastatic cells.
According to the scoring results of pathologists, the immunohistochemical staining results of patients were divided into a negative group and positive group. As shown in Figure 1D, the normal mucosal epithelium was strongly positive for ANXA1 expression, while both positive expression levels and a lack of ANXA1 expression were found in tumor tissues. To determine the role of ANXA1 expression in LN metastasis of HSCC, we evaluated the expression of ANXA1 in patients with and without LN metastasis by immunohistochemistry. About 37.5% (9/24) of patients without LN metastasis showed negative staining for ANXA1. However, ANXA1 staining was negative in 82% (41/50) of patients with LN metastasis. The results showed that the expression of ANXA1 was significantly lower in patients with LN metastasis than in those without LN metastasis (Table 2, p<0.05). Next, we analyzed the correlation between ANXA1 expression and clinicopathological features in 74 patients with HSCC. As shown in Table 3, the expression of ANXA1 was related to lymph node stage (p<0.05) and pathological differentiation (p<0.05). However, the expression of ANXA1 was not related to gender, age and tumor stage. Furthermore, our overall survival time analysis showed that patients with low expression of ANXA1 had a poor overall survival (hazard ratio [HR] =3.597; 95% confidence interval [CI], 1.410–9.176, p<0.05) (Figure 1E). These data suggested that ANXA1 plays a vital role in LN metastasis of HSCC.
 |
Table 2 The Immunohistochemical Expression of ANXA1 in HSCC |
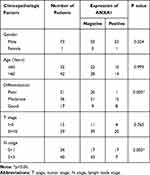 |
Table 3 Relationship Between ANXA1 Expression and Clinicopathological Features of HSCC |
Down-Regulation of ANXA1 Enhances Proliferation, Migration, and Invasive Behavior of FaDu Cells
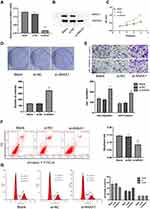
We then went on to investigate the biological function of ANXA1 in FaDu cells via ANXA1 silencing. First, we used qRT-PCR and Western blotting to analyze the ANXA1 mRNA and protein expression in FaDu cells transfected with si-NC or si-ANXA1. As expected, the levels of ANXA1 mRNA were significantly decreased in FaDu cells transduced with si-ANXA1 (p<0.0001) compared with the si-NC group (Figure 2A). Additionally, Western blotting showed that ANXA1 protein expression was significantly decreased in the si-ANXA1 group compared with the si-NC group (Figure 2B). These results confirmed that our siRNA approach successfully inhibited the expression of ANXA1 in FaDu cells.
We then studied the effects of ANXA1 on the proliferation, invasion and migration of FaDu cells by CCK-8 staining, colony formation and Transwell assays. As shown in Figure 2C–E, the si-NC group did not affect the growth, invasion or migration of FaDu cells. Conversely, compared with the si-NC group, the proliferative, invasive and migratory abilities of FaDu cells were significantly enhanced upon ANXA1 silencing. In addition, we assessed apoptosis levels and cell cycle status by FCM. As presented in Figure 2F, there was no difference in the apoptosis rate between the blank group and the si-NC group, while ANXA1 silencing significantly reduced the apoptosis rate compared with the si-NC group (p<0.01). Moreover, compared with the si-NC group, the proportion cells in the G0/G1 phase cells were significantly decreased in the si-ANXA1 group (p<0.05), while the proportion of cells in the S phase cells was significantly increased (p<0.05; Figure 2G). The results suggested that ANXA1 silencing may promote proliferative and invasive behaviors of cancer cells.
ANXA1 Expression is Closely Related to Yap1 and Plays an Important Role in HSCC LN Metastasis
In order to understand the molecular mechanisms underlying the role of ANXA1 in HSCC LN metastasis, we carried out an in-depth exploration of the RNA sequencing results. GO terms of biological processes (BP) related to ANXA1 were identified (Figure 3A) and demonstrated that ANXA1 was mainly involved in epithelial cell differentiation, regulation of cell proliferation and negative regulation of the apoptotic process (biological processes associated with cancer). The KEGG pathway enrichment results of all differentially expressed mRNA are shown in Figure 3B. The Hippo pathway was highly enriched, indicating that this pathway may play a key role in LN metastasis of HSCC. Hippo is one of eight major signal transduction pathways in human cancer, and Yap1 is a key factor in this pathway.22 Further analysis showed that Yap1 was involved in most of the same biological processes as ANXA1 (Figure 3C). Protein–protein interaction (PPI) networks of genes involved in the cancer biological processes with ANXA1 were constructed using the STRING database. This revealed that both ANXA1 and Yap1 were central genes in these pathways and were closely related to each other (Figure 3D).
Down-Regulation of ANXA1 Decreases Yap1 Protein Levels
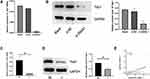
Hannah et al demonstrated that Yap1 was the most down-regulated protein in ANXA1−/− cells compared with ANXA1+/− cells.11 In order to explore the relationship between ANXA1 and Yap1, we assessed the mRNA and protein expression levels of Yap1 in FaDu cells (control group, si-NC group and si-ANXA1 group) and found that both Yap1 mRNA levels and protein levels were significantly reduced after ANXA1 silencing (p<0.01 and p<0.05, respectively; Figure 4A and B). We furthermore assessed Yap1 mRNA and protein levels in tissues of HSCC with or without LN metastasis and found that Yap1 mRNA was significantly lower in the LN metastasis group (p<0.01; Figure 4C), which was consistent with the RNA sequencing results. Moreover, the Yap1 protein expression levels were also significantly decreased in HSCC with LN metastasis (p<0.05; Figure 4D). Pearson correlation analysis showed that ANXA1 was positively correlated with Yap1 in HSCC tissue (p<0.0001; Figure 4E). Based on bioinformatic methods and tissue expression analysis, we therefore suggested that there was a correlation between ANXA1 and Yap1, and ANXA1 may regulate Yap1 expression in LN metastasis tissues of patients with HSCC.
Over-Expression of Yap1 Reverses the Effect of ANXA1 Silencing
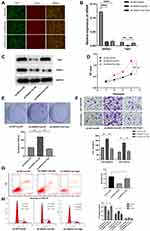
Based on the regulatory effect of ANXA1 on Yap1, we further verified the effect of Yap1 over-expression on the function of FaDu cells after ANXA1 silencing. First, we used lentivirus to construct sh-NC+oe-NC (negative control (NC) group), sh-ANXA1+oe-NC (ANXA1-silenced group) and sh-ANXA1+oe-Yap1(ANXA1-silenced/Yap1-overexpressed group) FaDu cell lines. In order to verify the efficiency of lentivirus transfection in FaDu cells, GFP and Cherry expression were examined using fluorescence microscopy in the three experimental groups (Figure 5A), and our results showed that the transfection efficiency was higher than 90%. We further used qRT-PCR and Western blotting to quantify the mRNA and protein expression of ANXA1 and Yap1 in the three groups (Figure 5B and C). The mRNA levels of ANXA1 in ANXA1-silenced group and ANXA1-silenced/Yap1-overexpressed group were significantly decreased (both p<0.0001), and the mRNA levels of Yap1 in ANXA1-silenced group were also significantly decreased (p<0.01) compared with their respective NC groups. Meanwhile, the mRNA levels of Yap1 in ANXA1-silenced/Yap1-overexpressed group were significantly higher than ANXA1-silenced group (p<0.01). In addition, Western blotting showed that the expression of ANXA1 was significantly decreased in the ANXA1-silenced group and ANXA1-silenced/Yap1-overexpressed group compared with their respective NC group. Likewise, the expression of Yap1 in the ANXA1-silenced group was significantly decreased compared with the NC group, but Yap1 expression was significantly increased in the ANXA1-silenced/Yap1-overexpressed group compared with the ANXA1-silenced group. These results indicated that sh-ANXA1 effectively inhibited the expression of ANXA1 in FaDu cells, and oe-Yap1 successfully overexpressed the expression of Yap1. At the same time, these experiments further showed that silencing ANXA1 reduced Yap1 expression.
As shown in Figure 5D–F, the results of CCK8 staining, colony formation and Transwell assays showed that ANXA1 silencing significantly enhanced cell activity, proliferation, migration or invasion compared with the NC group, while over-expression of Yap1 reversed the effect of ANXA1 silencing in FaDu cells. As shown in Figure 5G, the apoptosis of the ANXA1-silenced group was significantly reduced (P<0.05) compared with the NC group, but the apoptosis of FaDu cells was significantly increased after over-expression of Yap1 (P<0.05) compared with the ANXA1-silenced group. The proportion of cells in G0/G1 phase was significantly decreased (P<0.05) and the proportion of cells in S phase was significantly increased (P<0.05) in the ANXA1-silenced group compared with the NC group, while the proportion of cells in G0/G1 phase was significantly increased (P<0.05) and the proportion of cells in S phase was significantly decreased (P<0.05) compared with the ANXA1-silenced group after over-expression of Yap1 (Figure 5H). These results suggested that over-expression of Yap1 can rescue the effect of ANXA1 silencing.
Discussion
Due to its concealed location, most HSCC patients have cervical LN metastasis already at initial treatment.1–3 LN metastases considerably affect the prognosis of patients and are main cause of treatment failure.4–9,23 Previously, our team has applied RNA sequencing to assess differentially expressed genes in HSCC patients with or without LN metastasis and identified ANXA1 as one of the differentially expressed genes. Here, we show that ANXA1 may play a potentially important role in HSCC LN metastasis.
Several studies have shown that down-regulation of ANXA1 is involved in the occurrence, invasion, metastasis and progression of a variety of cancers.18,24,25 Our present analysis shows that the significant down-regulation of ANXA1 in HSCC patients with LN metastasis was closely associated with several clinicopathological features, such as poor cell differentiation and late stages of LN metastasis. In addition, patients with lower ANXA1 expression levels had poor survival. ANXA1 may therefore not only be useful for the future development of early diagnosis markers of LN metastasis in patients with HSCC, but may also represent as a prognostic factor for survival of HSCC patients. We used siRNA and sh-ANXA1 to silence the expression of ANXA1 in FaDu cells to explore the effect of ANXA1 on cell function. Down-regulation of ANXA1 in FaDu cells promoted their proliferation, metastasis and invasion and furthermore increased the proportion of cells in the S phase and decreased apoptosis levels.
Yap1 is the primary downstream effector of Hippo signaling pathway.26 Yap1 transfers from the cytoplasm to the nucleus, interconnects with promoters of various specific transcription factors, and plays a role in transcriptional activity, thus not only regulating growth of normal organs, but is also associated with tumorigenesis, tumor development, tumor metastasis and chemoresistance. Although there have been many recent reports on Yap1 in the area of tumor research, findings relating to its impact on the occurrence and development of tumors are inconsistent. A previous study has suggested that Yap1 expression has no significant correlation with the volume of schwannomas.27 Yap1 has also been confirmed as oncogene in many epithelial cells.26 While several studies have conversely revealed that Yap1 may represent a tumor suppressor in some tumor types and can promote apoptosis.28,29 A plausible explanation for this differential function of Yap1 is the environment in which the cells are located.30 Cells of large organisms are constantly damaged by various internal and external sources, which activates the DNA damage response (DDR).31 Yap1/Hippo is a key protector of genomic integrity in response to DNA damage which can respond to DNA damage by interacting with tumor suppressor genes p53 and p73 and subsequently regulates cell proliferation and apoptosis.32 However, if the DDR does not work properly, the DNA damage is not resolved and this can trigger genomic instability, enhancing the malignant growth of tumor cells. Using bioinformatic tools, we found that ANXA1 and Yap1 may be closely related and play an important role in HSCC with LN metastasis. Our in vitro experiments confirmed that ANXA1 regulates the expression of Yap1 in FaDu cells. Moreover, correlation analysis of mRNA expression levels of ANXA1 and Yap1 in HSCC tissues revealed that they are positively correlated. Over-expression of Yap1 could rescue the effect of ANXA1 silencing. Therefore, we can reasonably speculate that a decrease of Yap1 expression may lead to an imbalance of the DNA damage response in FaDu cells, and eventually result in LN metastasis of HSCC. However, further studies are needed to confirm our hypothesis.
The main strengths of this study were: (1) mRNA sequencing was performed for HSCC with and without LN metastasis; (2) we verified the expression of ANXA1 based on mRNA, protein and immunohistochemistry; (3) taking the low incidence of HSCC into consideration, the sample size of our immunohistochemistry analyses was relatively large; (4) we clarified the relationship between ANXA1 and Yap1 through a combination of bioinformatics analysis, functional in vitro experiments and quantification in tissue samples; (5) our judgments of surgical treatment, surgical procedures, and patient follow-up were conducted by a team of researchers rather than only one individual. The main weaknesses of the present study are that the overall number of specimens is limited and we also need to further explore the underlying molecular mechanism in more depth, which will be a focus of our next work.
Taken together, our study demonstrates that the expression of ANXA1 is decreased in HSCC tissues from patients with LN metastasis. Moreover, the survival rate of patients with low ANXA1 expression is reduced compared with patients with high ANXA1 expression. Therefore, in the future clinical work, we hope to use ANXA1 as a tumor marker to predict the LN metastasis and prognosis of HSCC patients. In our preliminary study investigating the mechanisms of ANXA1 inhibition of HSCC with LN metastasis, we found that ANXA1 may affect the LN metastasis by regulation of Yap1 expression. This provides a potential target for treatment of HSCC with LN metastasis.
Conclusion
In conclusion, this study investigated the expression and functional role of ANXA1 in LN metastasis of HSCC. The ANXA1/Yap1 axis may represent a potential target and a new theoretical basis for the treatment of HSCC with LN metastasis.
Acknowledgments
We would like to thank the center for molecular medicine and cancer research of Chongqing Medical University for providing experimental equipment and experimental platform. Thanks for the technical support from the Department of Pathology, the First Affiliated Hospital of Chongqing Medical University. This study was supported by the National Natural Science Foundation of China (Grant No. 81902776) and the Chongqing Joint Project of Science and Health (Grant No. 2020MSXM091).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu WZ, Li ZD, Li SC, et al. Study on the metastatic sequence of cervix lymph node in hypopharyngeal carcinoma. Zhonghua Zhong Liu Za Zhi. 2017;39(12):937–941. doi:10.3760/cma.j.issn.0253-3766.2017.12.011
2. Wang H, Wu R, Huang X, et al. The pattern of cervical lymph node metastasis and risk factors of retropharyngeal lymph node metastasis based on magnetic resonance imaging in different sites of hypopharyngeal carcinoma. Cancer Manag Res. 2020;12:8581–8587. doi:10.2147/CMAR.S245988
3. Buckley JG, MacLennan K. Cervical node metastases in laryngeal and hypopharyngeal cancer: a prospective analysis of prevalence and distribution. Head Neck. 2000;22(4):380–385. doi:10.1002/1097-0347(200007)22:4<380::AID-HED11>3.0.CO;2-E
4. Cooper JS, Porter K, Mallin K, et al. National cancer database report on cancer of the head and neck: 10-year update. Head Neck. 2009;31(6):748–758. doi:10.1002/hed.21022
5. Ahn D, Kim JH, Sohn JH, Sin CM, Lee JE. Laryngeal preservation in stage III/IV resectable laryngo-hypopharyngeal squamous cell carcinoma following concurrent chemoradiotherapy with capecitabine/cisplatin. Mol Clin Oncol. 2013;1(4):685–691. doi:10.3892/mco.2013.113
6. Li M, Lorenz RR, Khan MJ, et al. Salvage laryngectomy in patients with recurrent laryngeal cancer in the setting of nonoperative treatment failure. Otolaryngol Head Neck Surg. 2013;149(2):245–251. doi:10.1177/0194599813486257
7. Zhan F, Barlogie B, Arzoumanian V, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109(4):1692–1700. doi:10.1182/blood-2006-07-037077
8. Spector JG, Sessions DG, Haughey BH, et al. Delayed regional metastases, distant metastases, and second primary malignancies in squamous cell carcinomas of the larynx and hypopharynx. Laryngoscope. 2001;111(6):1079–1087.
9. Roberts TJ, Colevas AD, Hara W, Holsinger FC, Oakley-Girvan I, Divi V. Number of positive nodes is superior to the lymph node ratio and American joint committee on cancer N staging for the prognosis of surgically treated head and neck squamous cell carcinomas. Cancer. 2016;122(9):1388–1397.
10. Gobbetti T, Cooray SN. Annexin A1 and resolution of inflammation: tissue repairing properties and signalling signature. Biol Chem. 2016;397(10):981–993. doi:10.1515/hsz-2016-0200
11. Swa HL, Blackstock WP, Lim LH, Gunaratne J. Quantitative proteomics profiling of murine mammary gland cells unravels impact of annexin-1 on DNA damage response, cell adhesion, and migration. Mol Cell Proteomics. 2012;11(8):381–393. doi:10.1074/mcp.M111.011205
12. Huang Y, Zhang C, Chen C, et al. Investigation of circulating antibodies to ANXA1 in breast cancer. Tumour Biol. 2015;36(2):1233–1236. doi:10.1007/s13277-014-2751-x
13. Lin Y, Lin G, Fang W, Zhu H, Chu K. Increased expression of annexin A1 predicts poor prognosis in human hepatocellular carcinoma and enhances cell malignant phenotype. Med Oncol. 2014;31(12):327. doi:10.1007/s12032-014-0327-7
14. Roth U, Razawi H, Hommer J, et al. Differential expression proteomics of human colorectal cancer based on a syngeneic cellular model for the progression of adenoma to carcinoma. Proteomics. 2010;10(2):194–202. doi:10.1002/pmic.200900614
15. Moghanibashi M, Jazii FR, Soheili ZS, et al. Proteomics of a new esophageal cancer cell line established from Persian patient. Gene. 2012;500(1):124–133. doi:10.1016/j.gene.2012.03.038
16. Paweletz CP, Ornstein DK, Roth MJ, et al. Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60(22):6293–6297.
17. Faria PC, Sena AA, Nascimento R, et al. Expression of annexin A1 mRNA in peripheral blood from oral squamous cell carcinoma patients. Oral Oncol. 2010;46(1):25–30. doi:10.1016/j.oraloncology.2009.09.003
18. Cheng AL, Huang WG, Chen ZC, et al. Identification of novel nasopharyngeal carcinoma biomarkers by laser capture microdissection and proteomic analysis. Clin Cancer Res. 2008;14(2):435–445. doi:10.1158/1078-0432.CCR-07-1215
19. Xia J, Benner MJ, Hancock RE. NetworkAnalyst–integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42(WebServer issue):W167–174. doi:10.1093/nar/gku443
20. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
21. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
22. Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337e310.
23. Li Y, Lu T, Hu G. Gene sequencing and expression of Raf-1 in lymphatic metastasis of hypopharyngeal carcinoma. Cancer Biomark. 2020;28(2):181–191. doi:10.3233/CBM-191238
24. Jorge YC, Mataruco MM, Araujo LP, et al. Expression of annexin-A1 and galectin-1 anti-inflammatory proteins and mRNA in chronic gastritis and gastric cancer. Mediators Inflamm. 2013;2013:152860. doi:10.1155/2013/152860
25. Guo C, Liu S, Sun MZ. Potential role of Anxa1 in cancer. Future Oncol. 2013;9(11):1773–1793. doi:10.2217/fon.13.114
26. Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi:10.1016/j.devcel.2010.09.011
27. Alessandro M, Gino M, Elisabetta Z, et al. Yap, Taz and Areg expression in eighth cranial nerve schwannoma. Int J Biol Markers. 2017;32(3):e319–e324. doi:10.5301/ijbm.5000263
28. Yuan M, Tomlinson V, Lara R, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15(11):1752–1759. doi:10.1038/cdd.2008.108
29. Cottini F, Hideshima T, Xu C, et al. Rescue of hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20(6):599–606. doi:10.1038/nm.3562
30. Bertini E, Oka T, Sudol M, et al. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8(1):49–57. doi:10.4161/cc.8.1.7259
31. Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc Natl Acad Sci U S A. 2000;97(2):686–691. doi:10.1073/pnas.97.2.686
32. Raj N, Bam R. Reciprocal crosstalk between YAP1/hippo pathway and the p53 family proteins: mechanisms and outcomes in cancer. Front Cell Dev Biol. 2019;7:159. doi:10.3389/fcell.2019.00159
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.