Back to Journals » International Journal of General Medicine » Volume 15
Expression and Clinical Values of Serum miR-155 and miR-224 in Chinese Patients with HCV Infection
Authors Jin X, Zhang Y, Wang H, Zhang Y
Received 19 October 2021
Accepted for publication 19 January 2022
Published 10 February 2022 Volume 2022:15 Pages 1393—1403
DOI https://doi.org/10.2147/IJGM.S344345
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Xiaochun Jin,1,* Ying Zhang,1,* Hui Wang,2 Youtao Zhang3
1Department of Anesthesiology, Suzhou Kowloon Hospital, Shanghai Jiaotong University School of Medicine, Suzhou, Jiangsu, People’s Republic of China; 2Department of Neurology, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China; 3Center of Clinical Laboratory, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Youtao Zhang, Center of Clinical Laboratory, First Affiliated Hospital of Soochow University, 188 Shizi Road, Suzhou, 215000, People’s Republic of China, Tel/Fax +8617710114047, Email [email protected] Hui Wang, Department of Neurology, First Affiliated Hospital of Soochow University, 188 Shizi Road, Suzhou, 215000, People’s Republic of China, Tel/Fax +8613913594769, Email [email protected]
Background: To investigate the expression of serum miR-155 and miR-224 among patients with hepatitis C virus (HCV) infection and analyze their clinical values.
Methods: A total of 116 patients suffering from HCV infection admitted to our hospital and 70 healthy subjects were selected. According to the diagnostic results, patients with HCV infection were divided into 48 cases of chronic hepatitis C (CHC), 43 cases of liver cirrhosis and 25 cases of hepatocellular carcinoma (HCC). The expression signature for miR-155 and miR-224 was detected in serum samples. ROC curve and Pearson correlation test were conducted to investigate their diagnostic value and correlation.
Results: The expression extent for serum miR-155 and miR-224 increased along with the increase of malignancy (all P < 0.05). According to ROC curve, the area under the curve (0.918, 95% CI: 0.856– 0.974) of miR-155 and miR-224 combined in the diagnosis of HCC was the largest, and its sensitivity and specificity were 93.0% and 86.2%. There is a positive relationship for expression level between miR-155 and miR-224 in CHC and HCC group (all P < 0.001).
Conclusion: miR-155 and miR-224 are remarkably increased in patients suffering from HCV infection. The combination of miR-155 and miR-224 has a good diagnostic value for HCC caused by HCV infection.
Keywords: hepatitis C, hepatocellular carcinoma, miR-155, miR-224
Introduction
As a severe health issue across the world, hepatitis C virus (HCV) infection is expected to affect 1% of the global population, and the majority is not aware until the growth of advanced liver disease.1 Nowadays, 400,000 people die of HCV infection each year. It is predicted that by 2040, more than 800,000 cases are expected to die from HCV-associated cirrhosis or HCC.2,3 HCV can trigger chronic hepatitis C (CHC), which is also one of the main causes of HCC. In China, chronic HCV infection is considered to be a very important factor for cirrhosis, chronic hepatitis and HCC. According to epidemiological studies conducted in the mid-1990s reported a national prevalence of 3.2%,4 but other reports suggest a range of 0.29% to 9.6%.4 The incidence of cirrhosis is 5% to 25% after 25 to 30 years of HCV infection,5 liver decompensation is 30% after 10 years of HCV-associated cirrhosis,5 and hepatocellular carcinoma is 1% to 3% per year.5,6 Chronic HCV infection is one of the most common risk factors for HCC. It is estimated that there are about 10 million HCV patients in China. Chronic hepatitis mainly causes extensive fibrosis and HCC through chronic inflammation, cell necrosis and regeneration.
There is currently no vaccine to prevent HCV infection. The combination of long-acting pegylated interferon type I (PEGIFN-α) and ribavirin (RBV) is the optimal treatment for hepatitis C in recent 10 years, but more than 50% of the patients with type I HCV still do not respond to this combination of antiviral therapy. At the same time, there are obvious toxic and side effects (such as influenza-like symptoms, anemia and depression, etc), and the treatment course is long, usually up to 1 year, so the clinical application of this therapy is limited. According to the current situation, antiviral therapy for chronic HCV infection has entered the era of direct antiviral drug pangenotype. A generic interferon-free regimen is preferred, which achieves a sustained virological response of more than 90% in HCV-infected patients with known major genotypes and major subtypes, and is consistent in multiple populations with different clinical characteristics and less drug interactions. Ribavirin (RBV) is not required in combination with ribavirin (RBV), except in a few specific populations, such as decompensated cirrhosis and DAAs failure. Therefore, the use of a pan-genotype regimen reduces pretreatment testing and in-treatment monitoring and is more suitable for the treatment and management of patients with chronic HCV infection at the primary level. Therefore, finding a reliable biomarker for the diagnosis of HCV-associated disease is of great importance.
MicroRNA (miRNA), is a cluster of about 21–25 nucleotides, which regulates gene expression through targeting mRNA. miRNA is small, conserved, noncoding and endogenous. miRNA is involved in a variety of biological and pathological processes such as cell development, signal transduction, cell growth, proliferation, movement and apoptosis.7,8 As a small-molecule RNA, is abnormally expressed in patients suffering from hepatitis C, participating in the occurrence and development of hepatitis C as a hidden target for hepatitis C treatment.9,10 Multiple miRNAs are dysregulating and have been recognized for being potential and promising biomarkers in HCC diagnosis and staging,11,12 including miR-30b,13 miR-485,14 miR-545,15 miR-30a-5p,16 miR-214,17 miR-497-5p18 and miR-9-5p.19 According to current researches, miR-155 and miR-224 participate in mediating hepatocyte immune response, exert a significant effect on the progression and metastasis of HCC, and are expected to become potential markers for the diagnosis of HCC.20,21 This study analyzed the expression of miR-155 and miR-224 among patients suffering from HCV-infected liver disease, so as to explore their diagnostic values for HCC and to provide guidance for diagnosing and treating HCC.
Materials and Methods
Research Objects
In total, 116 patients with HCV infection receiving treatment in our hospital from May 1, 2020 to May 31, 2021 were collected as the research objects, including 62 males and 54 females, with an average of (47.50±9.82) years. These patients were fallen into three groups including 48 cases of chronic hepatitis C, 43 cases of liver cirrhosis and 25 cases of HCC. The 116 patients were infected with HCV, including 76 cases of type 1B, 36 cases of type 2A, 3 cases of type 3A, and 1 case of type 3B. TNM stages of 25 HCC patients included stage I–II in 12 cases and stage III–IV in 13 cases. In this study, direct antiviral drugs were used to treat patients infected with HCV in accordance with the latest WHO guidelines for screening and prevention of chronic hepatitis C released in 2016. The antiviral drugs were mainly sofebuvir combined with daclatasvir, and oral antiviral drugs of GI II and GI III. Another 70 cases undergoing physical examination were enrolled as controls including 39 males and 31 females, with an average of (48.73±8.95) years.
Inclusion and Exclusion Criteria
Inclusion criteria were as follows: 1) diagnosis of CHC refers to the criteria in the guidelines for preventing and treating hepatitis C22; 2) HCV RNA was quantitatively positive in liver tissue. Exclusion criteria were as follows: 1) patients with liver disease induced by drug, fatty liver disease and hereditary metabolic liver disease; 2) serological examination combined with other types of hepatitis virus infection, EB virus, cytomegalovirus, HIV-infected persons, malignant tumors and immune diseases; 3) patients with severe heart, brain and kidney diseases; 4) HCV-infected persons who are receiving or have previously received interferon or interferon combined with ribavirin antiviral therapy.
Detection Indexes
The biochemical data measured and how they were done we consult the corresponding references.23,24 HCV-RNA detection: all subjects used SLAM96P fluorescence quantitative PCR instrument and reagent provided by Shanghai Hongshi Company to detect HCV-RNA before treatment. And we quantified produced RNA using A260/A280 ratio.25–27 Detection of miR-155 and miR-224: miR-155 and miR-224 were detected by SLAM96P fluorescence quantitative PCR (Thermo Scientific, Waltham, Massachusetts, USA). The kit of RNA extraction: Invitrogen TRIzol (Thermo Scientific, Waltham, Massachusetts, USA). The total PCR system was 20 mL: 10 mL TaqMan general mixture solution (2×), 1 mL primer and probe Mix (20 ×), 1.33 mL reverse transcribed DNA, 7.67 mL double distilled water. The detailed experimental protocols and methods for total RNA extraction, cDNA synthesis, qRT-PCR and biochemical tests can be consulted in the literature by Chen et al.28,29 Forward primer: 5’-GCCGCTTAATGCTAATCGTG-3, reverse primer: 5’- CAGTGCTGGGTCCGACTGA-3’ for miR-155 gene, forward primer: 5’-CAAGUCACUAGUGGUUCCGUU-3’, reverse primer: 5’- TCCACCACTGTCCCGACT -3’ for miR-224 gene and U6 gene was used as an internal control using the forward primer: 5’-CTCGCTTCGGCAGCACA-3’ and reverse primer: 5’-CAAGTCACTAGTGGTTCCGTTTA-3’. We used the 2−ΔΔCt calculation formula to determine the relative expression level as follows: ΔCt=CT value of target gene (miR-155 or miR-224)-CT value of U6.30–33
Statistical Methods
The statistical analyses were conducted through SPSS20.0 software. x±S was employed to represent measurement data exhibiting a normal distribution. Comparisons between groups were analyzed with the independent sample t-test. Enumeration data between groups was compared with the χ2 test. Single-factor survival analysis and multivariate survival analysis were conducted by employing Kaplan–Meier, Log rank and Cox regression analyses. P<0.05 was considered to be statistically significant.
Ethical Approval/Patient Consent
The present study was approved by the Ethics Committee of first affiliated hospital of Soochow University (number: 129/2020, dated 19/04/2020). All patients, control subjects, and their family members provided informed consent. All materials and methods conformed to correlative policies and regulations. This investigation was performed in accordance with the declaration of Helsinki.
Results
Clinical Data
The features of research population are shown in Table 1. Three groups showed greatly higher extents of ALT and AST than control group (P<0.01). Other characteristics including age, gender, BMI or HCV-RNA content displayed no great differences. There was no significance in parameters including age, gender, BMI or HCV-RNA content. Compared with control group, significantly higher extents of ALT and AST were detected in CHC, liver cirrhosis and hepatocellular carcinoma.
 |
Table 1 The Characteristic of Study Population |
Expression Levels of miR-155 and miR-224
The expression extents of serum miR-155 and miR-224 increased along with the increase of malignancy (all P<0.05). The detailed information is shown in Table 2.
 |
Table 2 miR-155 and miR-224 Expression Level in Different Groups |
 |
Table 3 Serum miR-155 and miR-224 Cutoff Points and the Corresponding Sensitivity and Specificity for Predicting HCC |
For both miR-155 and miR-224, gene expression is highest in HCC patients, followed by cirrhosis and HCV patients, and lowest in healthy people.
The Values of miR-155 and miR-224 for HCC Diagnosis
The area under the curve of miR-155 and miR-224 in the combined diagnosis of HCC was higher than that of single miR-155 and miR-224, the diversity was of statistical significance (z=4.107, 4.315, P=0.016, 0.005), the sensitivity was 93.0%, and the specificity was 86.2%. There was no significant difference in the region under the curve of miR-155 in the diagnosis of HCC compared with miR-224 (z=1.108, P = 0.217). The detailed information was shown in Table 3 and Figure 1.
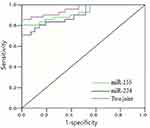 |
Figure 1 ROC curve of miR-155 and miR-224 combined in the diagnosis of HCC. Abbreviation: HCC, hepatocellular carcinoma. |
Relationship of Expression Level Between miR-155 and miR-224
According to relationship analysis, the expression level of miR-155 was positively related to miR-224 in CHC group (r=0.716, P<0.001). The similar phenomenon was also occurred in miR-155 and miR-224 in HCC group (r=0.784, P<0.001). The detailed information was shown in Figures 2–4.
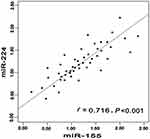 |
Figure 2 Correlation between miR-155 expression level and miR-224 in CHC. Abbreviation: CHC, chronic hepatitis C. |
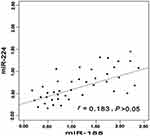 |
Figure 3 Correlation between miR-155 expression level and miR-224 in liver cirrhosis. |
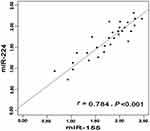 |
Figure 4 Correlation between miR-155 expression level and miR-224 in HCC. Abbreviation: HCC, hepatocellular carcinoma. |
Discussion
CHC is the most important cause of end-stage liver disease in Europe, the United States and Japan, according to the World Health Organization statistics, there are more than 170 million patients infected with HCV worldwide.34,35 Cirrhosis due to chronic HCV infection is the primary index for liver transplantation and a major factor in the increased incidence of HCC.7,8,36–38
MiRNA is a cluster of about 21–25 nucleotides, which regulates gene expression through targeting mRNA. miRNA is small, conserved, noncoding and endogenous. miRNA is involved in a variety of biological and pathological processes such as cell development, signal transduction, cell growth, proliferation, movement and apoptosis.7,8,34 It is common that miRNAs is expressed abnormally. miRNA is a kind of endogenous non-coding single stranded small molecules with a length of about found in eukaryotes in recent years. It can combine with the corresponding target genes through base pairing, resulting in target degradation or post-transcriptional translation inhibition, and control gene expression at the post-transcriptional extent. Studies have shown that it not only takes part in a lot of biological processes including cell division, proliferation, differentiation and development but also plays a function similar to oncogenes and tumor suppressor genes, and exerts a significant effect on tumor cell proliferation, apoptosis, differentiation and metabolism.39–44 MiRNAs regulate and control the formation and development of tumors by interacting with various signaling pathways such as EGFR/STAT3, JAK2/STAT3, NF-κB, AKT/mTOR. Recently, several possible key player targets in cancer that have been linked to various cancer types such as Akt, Topo-isomerase II-B, CTLA4, STAT4, FXR.45–48
MiRNA exerts a significant effect on the development of CHC, hepatocyte regeneration and functional regulation as a hidden marker for diagnosing and treating HCC. miR-155 and miR-224 have been extensively studied in various types of diseases and multiple countries and regions, especially in China. It has been found that aberrant expression of miR-155 displays as an oncogenic feature in various types of hematological malignancies and solid tumors.49–52 Furthermore, it has been studied in multiple inflammatory diseases including asthma,53 sepsis,54 periodontitis55 and rheumatoid arthritis.56 miR-224 regulates the expression of upstream and downstream genes and affects the occurrence and progression of diseases including liver cancer,57 colorectal cancer,58 uveal melanoma59 and cervical cancer.60 Zhang et al have shown that miR-155 participates in developing and differentiating immune cells with high expression in HCC tissues.61 It can promote the appearance, invasion and metastasis of HCC cells, and exerts a significant effect on the prognosis and targeted treatment of HCC. Cui et al found that miR-224 is the promoter of HCC, and its expression in hepatocellular carcinoma is related to tumor cell propagation, migration, aggression and prognosis, which can be used as a diagnostic and prognostic index of HCC.62
According to this study, by comparing with the control group, miR-155 and miR-224 were remarkably increased in HCC group, liver cirrhosis group and CHC group, and the HCC group was noticeably higher than CHC group and liver cirrhosis group. To the best of our knowledge, the relationship between miR-155 and CHC infection has been reported in only a few literatures in Caucasian population. And the relationship between miR-224 and CHC infection has not been reported. It is suggested that the high expression of miR-155 and miR-224 may take part in occurring and developing hepatitis C. Some studies have found that the level of serum miR-224 in patients suffering from HCV-related hepatitis and HCC is related to the grade of hepatitis, the degree of liver fibrosis and the degree of tumor tissue differentiation.63,64 Monitoring the expression of miR-224 may be helpful to diagnosing and treating response evaluation of patients suffering from HCC. The ROC curve of this study indicates that the combination of miR-155 and miR-224 has the largest area under the curve for the diagnosis of HCC, with sensitivity of 93.0% and specificity of 86.2%. Correlation analysis also showed that miR-155 and miR-224 in HCC group are significantly positively correlated. This demonstrates that miR-155 and miR-224 may be biological indicators for diagnosing HCC, and the integrated detection of miR-155 and miR-224 has good values in the diagnosis of HCC. Researches have shown that the expression extent of miR-155 in HCC tissues and serum is greater than that in normal liver tissues and healthy controls.42 The region under the curve of miR-155 in diagnosing HCC is 0.840, which has high efficiency in the diagnosis of HCC and may be a good index for the diagnosis of HCC. Amr et al observed that the extent of miR-224 in HCC group was greatly higher than that in the CHC group and control group, which was associated with the appearance and growth of HCC.63 The expression of miR-224 could be adopted as a noninvasive biomarker for early prediction of HCC. According to other researches, the level of serum miR-224 in patients with early HCC is significantly higher than that in patients suffering from liver cirrhosis, chronic hepatitis and healthy controls.41,42 The area under the curve for identifying early HCC is 0.880. miR-224 is a reliable biomarker for the early detection of HCC. It should be noted that some useful molecular techniques used such as microarrays and RNA-Seq should be applied to detect miRNAs in the future.65–69
Conclusion
In summary, with the progress of the disease, the expression levels of serum miR-155 and miR-224 among patients suffering from hepatitis related to HCV infection increased strikingly. miR-155 and miR-224 are remarkably increased in patients suffering from HCV infection. The combination of miR-155 and miR-224 has a good diagnostic value for HCC caused by HCV infection.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Acknowledgments
We sincerely thank the graduate student named “Haitao Hu” for helping us in data collection and collation. Xiaochun Jin and Ying Zhang are co-first authors for this study.
Author Contributions
YZ conceived study design; XJ and YZ conceived the content concept; HW, XJ and YZ performed the data collection, extraction and analyzed the data. HW, XJ and YZ interpreted and reviewed the data and drafts. YZ and HW reviewed the final draft. All authors were involved in literature search, writing the paper and had final approval of the submitted and published versions. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Disclosure
The authors declare that they have no competing interests.
References
1. Ng L, Wan TM, Man JH, et al. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8:27393–27400. doi:10.18632/oncotarget.16171
2. Feng Y, Dong YW, Song YN, et al. MicroRNA449a is a potential predictor of colitis‑associated colorectal cancer progression. Oncol Rep. 2018;40:1684–1694. doi:10.3892/or.2018.6566
3. Zhang H, Wang Z, Ma R, Wu J, Feng J. MicroRNAs as biomarkers for the progression and prognosis of colon carcinoma. Int J Mol Med. 2018;42:2080–2088. doi:10.3892/ijmm.2018.3792
4. Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837–87847. doi:10.18632/oncotarget.21237
5. Liu X, Duan B, Dong Y, et al. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7:8046–8052.
6. Lohmann V, Bartenschlager R. Indelibly stamped by hepatitis C virus infection: persistent epigenetic signatures increasing liver cancer risk. Gastroenterology. 2019;156:2130–2133. doi:10.1053/j.gastro.2019.04.033
7. Saldanha J, Heath A, Aberham C, et al. World Health Organization collaborative study to establish a replacement WHO international standard for hepatitis C virus RNA nucleic acid amplification technology assays. Vox Sang. 2005;88:202–204. doi:10.1111/j.1423-0410.2005.00606.x
8. Freeman RB
9. Weis A, Marquart L, Calvopina DA, Genz B, Ramm GA, Skoien R. Serum microRNAs as biomarkers in hepatitis C: preliminary evidence of a microRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mol Sci. 2019;20:864.
10. Sadri Nahand J, Bokharaei-Salim F, Salmaninejad A, et al. microRNAs: key players in virus-associated hepatocellular carcinoma. J Cell Physiol. 2019;234:12188–12225. doi:10.1002/jcp.27956
11. El-Maraghy SA, Adel O, Zayed N, Yosry A, El-Nahaas SM, Gibriel AA. Circulatory miRNA-484, 524, 615 and 628 expression profiling in HCV mediated HCC among Egyptian patients; implications for diagnosis and staging of hepatic cirrhosis and fibrosis. J Adv Res. 2020;22:57–66. doi:10.1016/j.jare.2019.12.002
12. Yasser MB, Abdellatif M, Emad E, et al. Circulatory miR-221 & miR-542 expression profiles as potential molecular biomarkers in hepatitis C virus mediated liver cirrhosis and hepatocellular carcinoma. Virus Res. 2021;296:198341. doi:10.1016/j.virusres.2021.198341
13. Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268–1275. doi:10.1016/j.bbrc.2016.04.054
14. Liu Z, Dou C, Yao B, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. doi:10.18632/oncotarget.8129
15. Zhang XN, Zhou J, Lu XJ. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol. 2018;233:6733–6741. doi:10.1002/jcp.26371
16. Pan Y, Tong S, Cui R, et al. Long non-coding MALAT1 functions as a competing endogenous RNA to regulate vimentin expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell Physiol Biochem. 2018;50:108–120. doi:10.1159/000493962
17. Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2018;314:G559–G565. doi:10.1152/ajpgi.00242.2017
18. Zhang Y, Zhu Z, Huang S, et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 2019;19:198. doi:10.1186/s12935-019-0909-8
19. Liu Z, Chen JY, Zhong Y, Xie L, Li JS. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res. 2019;52:e8631. doi:10.1590/1414-431x20198631
20. Liao WW, Zhang C, Liu FR, Wang WJ. Effects of miR-155 on proliferation and apoptosis by regulating FoxO3a/BIM in liver cancer cell line HCCLM3. Eur Rev Med Pharmacol Sci. 2020;24:7196. doi:10.26355/eurrev_202007_21845
21. Wu C, Wang X, Zhang J, et al. MicroRNA-224 expression and polymorphism predict the prognosis of hepatitis B virus-related hepatocellular carcinoma patients after liver resection. Clin Lab. 2019;65. doi:10.7754/Clin.Lab.2018.181025
22. Chinese Society of Hepatology, Chinese Society of Infectious Diseases, Chinese Medical Association. [Guidelines for the prevention and treatment of hepatitis C (2019 version)]. Zhonghua Gan Zang Bing Za Zhi. 2019;27:962–979. Chinese. doi:10.3760/cma.j.issn.1007-3418.2019.12.008
23. Ebeid HM, Gibriel AA, Al-Sayed HM, Elbehairy SA, Motawe EH. Hepatoprotective and antioxidant effects of wheat, carrot, and mango as nutraceutical agents against CCl4-induced hepatocellular toxicity. J Am Coll Nutr. 2015;34:228–231. doi:10.1080/07315724.2014.887486
24. Ahmed SA, Gibriel AA, Abdellatif AK, Ebied HM. Evaluation of food products fortified with oyster shell for the prevention and treatment of osteoporosis. J Food Sci Technol. 2015;52:6816–6820. doi:10.1007/s13197-015-1725-3
25. Gibriel AA, Abou-Elew MH, Masmoudi S. Analysis of p.Gly12Valfs*2, p.Trp24* and p.Trp77Arg mutations in GJB2 and p.Arg81Gln variant in LRTOMT among non syndromic hearing loss Egyptian patients: implications for genetic diagnosis. Mol Biol Rep. 2019;46:2139–2145. doi:10.1007/s11033-019-04667-0
26. Gibriel AA, Tate RJ, Yu Y, et al. The p.Arg86Gln change in GARP2 (glutamic acid-rich protein-2) is a common West African-related polymorphism. Gene. 2013;515:155–158. doi:10.1016/j.gene.2012.11.005
27. Mosrati MA, Fadhlaoui-Zid K, Benammar-Elgaaied A, Gibriel AA, Ben Said M, Masmoudi S. Deep analysis of the LRTOMTc.242G>A variant in non-syndromic hearing loss North African patients and the Berber population: implications for genetic diagnosis and genealogical studies. Mol Genet Genomic Med. 2021;9:e1810. doi:10.1002/mgg3.1810
28. Chen X, Li P, Yang Z, Mo WN. Expression of fragile histidine triad (FHIT) and WW-domain oxidoreductase gene (WWOX) in nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 2013;14:165–171. doi:10.7314/APJCP.2013.14.1.165
29. Chen X, Zhang H, Li P, Yang Z, Qin L, Mo W. Gene expression of WWOX, FHIT and p73 in acute lymphoblastic leukemia. Oncol Lett. 2013;6:963–969. doi:10.3892/ol.2013.1514
30. Bouzid A, Smeti I, Dhouib L, et al. Down-expression of P2RX2, KCNQ5, ERBB3 and SOCS3 through DNA hypermethylation in elderly women with presbycusis. Biomarkers. 2018;23:347–356. doi:10.1080/1354750X.2018.1427795
31. Bouzid A, Smeti I, Chakroun A, et al. CDH23 methylation status and presbycusis risk in elderly women. Front Aging Neurosci. 2018;10:241. doi:10.3389/fnagi.2018.00241
32. Jiwaji M, Daly R, Gibriel A, et al. Unique reporter-based sensor platforms to monitor signalling in cells. PLoS One. 2012;7:e50521. doi:10.1371/journal.pone.0050521
33. Ben Ayed I, Ouarda W, Frikha F, et al. SRD5A3-CDG: 3D structure modeling, clinical spectrum, and computer-based dysmorphic facial recognition. Am J Med Genet A. 2021;185:1081–1090. doi:10.1002/ajmg.a.62065
34. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi:10.3322/caac.20107
35. Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi:10.3748/wjg.v16.i4.418
36. Roblero JP, Arab JP, Mezzano G, Mendizabal M. Hepatitis C virus infection: what are we currently doing in Latin America about WHO’s proposals for 2030? Clin Liv Dis. 2021;18:72–75. doi:10.1002/cld.1084
37. Foreman KJ, Marquez N, Dolgert A, et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet. 2018;392:2052–2090. doi:10.1016/S0140-6736(18)31694-5
38. Davila JA, Morgan RO, Shaib Y, McGlynn KA, El-Serag HB. Hepatitis C infection and the increasing incidence of hepatocellular carcinoma: a population-based study. Gastroenterology. 2004;127:1372–1380. doi:10.1053/j.gastro.2004.07.020
39. Mourad L, El-Ahwany E, Zoheiry M, et al. Expression analysis of liver-specific circulating microRNAs in HCV-induced hepatocellular carcinoma in Egyptian patients. Cancer Biol Ther. 2018;19:400–406. doi:10.1080/15384047.2018.1423922
40. Dai M, Li L, Qin X. Clinical value of miRNA-122 in the diagnosis and prognosis of various types of cancer. Oncol Lett. 2019;17:3919–3929. doi:10.3892/ol.2019.10024
41. Mamdouh S, Khorshed F, Aboushousha T, et al. Evaluation of Mir-224, Mir-215 and Mir-143 as serum biomarkers for HCV associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 2017;18:3167–3171. doi:10.22034/APJCP.2017.18.11.3167
42. Ning S, Liu H, Gao B, et al. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol Lett. 2019;18:3381–3387. doi:10.3892/ol.2019.10606
43. Zinkin NT, Grall F, Bhaskar K, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi:10.1158/1078-0432.CCR-07-0586
44. Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi:10.1016/j.ccr.2006.01.025
45. Abo-Salem HM, Gibriel AA, El Awady ME, Mandour AH. Synthesis, molecular docking and biological evaluation of novel flavone derivatives as potential anticancer agents targeting Akt. Med Chem. 2021;17:158–170. doi:10.2174/1573406416666200306115035
46. Mohamady S, Gibriel AA, Ahmed MS, Hendy MS, Naguib BH. Design and novel synthetic approach supported with molecular docking and biological evidence for naphthoquinone-hydrazinotriazolothiadiazine analogs as potential anticancer inhibiting topoisomerase-IIB. Bioorg Chem. 2020;96:103641. doi:10.1016/j.bioorg.2020.103641
47. Ali NA, Hamdy NM, Gibriel AA, El Mesallamy HO. Investigation of the relationship between CTLA4 and the tumor suppressor RASSF1A and the possible mediating role of STAT4 in a cohort of Egyptian patients infected with hepatitis C virus with and without hepatocellular carcinoma. Arch Virol. 2021;166:1643–1651. doi:10.1007/s00705-021-04981-8
48. Attia YM, Tawfiq RA, Gibriel AA, et al. Activation of FXR modulates SOCS3/Jak2/STAT3 signaling axis in a NASH-dependent hepatocellular carcinoma animal model. Biochem Pharmacol. 2021;186:114497. doi:10.1016/j.bcp.2021.114497
49. Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37:33–44. doi:10.1007/s10555-017-9724-7
50. Shao C, Yang F, Qin Z, Jing X, Shu Y, Shen H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: a systematic review with meta-analysis. BMC Cancer. 2019;19:1103. doi:10.1186/s12885-019-6297-6
51. Wang F, Xu W, Gu WY, Li JY. [Expression and prognostic value of MiR-155 in patients with chronic lymphocytic leukemia]. Zhongguo Shi Yan xue Ye Xue Za Zhi. 2020;28:34–39. Chinese. doi:10.19746/j.cnki.issn.1009-2137.2020.01.007
52. Cao H, Huang S, Liu A, Chen Z. Up-regulated expression of miR-155 in human colonic cancer. J Cancer Res Ther. 2018;14:604–607. doi:10.4103/0973-1482.175432
53. Karam RA, Abd Elrahman DM. Differential expression of miR-155 and Let-7a in the plasma of childhood asthma: potential biomarkers for diagnosis and severity. Clin Biochem. 2019;68:30–36. doi:10.1016/j.clinbiochem.2019.04.007
54. Lan C, Shi X, Guo N, Pei H, Zhang H. [Value of serum miR-155-5p and miR-133a-3p expression for the diagnosis and prognosis evaluation of sepsis]. Zhonghua wei Zhong Bing Ji Jiu Yi xue. 2016;28:694–698. Chinese. doi:10.3760/cma.j.issn.2095-4352.2016.08.005
55. Wu P, Feng J, Wang W. Expression of miR-155 and miR-146a in the saliva of patients with periodontitis and its clinical value. Am J Transl Res. 2021;13:6670–6677.
56. Abdul-Maksoud RS, Sediq AM, Kattaia A, et al. Serum miR-210 and miR-155 expression levels as novel biomarkers for rheumatoid arthritis diagnosis. Br J Biomed Sci. 2017;74:209–213. doi:10.1080/09674845.2017.1343545
57. Hung JH, Li CH, Yeh CH, et al. MicroRNA-224 down-regulates glycine N-methyltransferase gene expression in hepatocellular carcinoma. Sci Rep. 2018;8:12284. doi:10.1038/s41598-018-30682-5
58. Zheng Q, Yu JJ, Li C, Li J, Wang J, Wang S. miR-224 targets BTRC and promotes cell migration and invasion in colorectal cancer. 3 Biotech. 2020;10:485. doi:10.1007/s13205-020-02477-x
59. Li J, Liu X, Li C, Wang W. miR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma. J Cell Biochem. 2019;120:12412–12421. doi:10.1002/jcb.28507
60. Fang W, Shu S, Yongmei L, Endong Z, Lirong Y, Bei S. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci Rep. 2016;6:33229. doi:10.1038/srep33229
61. Zhang L, Wang W, Li X, et al. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol. 2016;48:2425–2434. doi:10.3892/ijo.2016.3465
62. Cui Y, Xu HF, Liu MY, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25:1890–1898. doi:10.3748/wjg.v25.i15.1890
63. Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215–221. doi:10.1016/j.gendis.2017.10.003
64. Lin L, Lu B, Yu J, Liu W, Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40:397–404. doi:10.1016/j.clinre.2015.11.005
65. Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi:10.1038/cr.2008.282
66. Gibriel AA. Options available for labelling nucleic acid samples in DNA microarray-based detection methods. Brief Funct Genomics. 2012;11:311–318. doi:10.1093/bfgp/els015
67. Andres-Leon E, Rojas AM. miARma-Seq, a comprehensive pipeline for the simultaneous study and integration of miRNA and mRNA expression data. Methods. 2019;152:31–40. doi:10.1016/j.ymeth.2018.09.002
68. Souissi A, Gibriel AA, Masmoudi S. Genetics and meta-analysis of recessive non-syndromic hearing impairment and Usher syndrome in Maghreb population: lessons from the past, contemporary actualities and future challenges. Hum Genet. 2021. doi:10.1007/s00439-021-02314-y
69. Souissi A, Ben Said M, Ben Ayed I, et al. Novel pathogenic mutations and further evidence for clinical relevance of genes and variants causing hearing impairment in Tunisian population. J Adv Res. 2021;31:13–24. doi:10.1016/j.jare.2021.01.005
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
