Back to Journals » International Journal of General Medicine » Volume 14
Expression and Clinical Significance of MPS-1 in Hepatocellular Carcinoma
Authors Jiang H, Yuan F, Zhao Z, Xue T, Ge N, Ren Z, Zhang L
Received 23 August 2021
Accepted for publication 1 November 2021
Published 1 December 2021 Volume 2021:14 Pages 9145—9152
DOI https://doi.org/10.2147/IJGM.S334378
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
He Jiang, Feifei Yuan, Zhiying Zhao, Tongchun Xue, Ningling Ge, Zhenggang Ren, Lan Zhang
Department of Hepatic Oncology, Zhongshan Hospital, Liver Cancer Institute and Key Laboratory of Carcinogenesis and Cancer Invasion, Fudan University, Shanghai, People’s Republic of China
Correspondence: Lan Zhang
Department of Hepatic Oncology, Zhongshan Hospital, Liver Cancer Institute and Key Laboratory of Carcinogenesis and Cancer Invasion, Fudan University, Shanghai, 200032, People’s Republic of China
Tel +86-021-64041990
Fax +86 021-64038472
Email [email protected]
Purpose: Ribosomal protein metallopanstimulin-1 (MPS-1) is implicated in tumorigenesis. However, to date, the underlying role of MPS-1 in the generation, progression and prognosis of hepatocellular carcinoma (HCC) remains unknown. This study aims to investigate the expression of MPS-1 in HCC and its significance for the prognosis of HCC.
Methods: The Oncomine and GEPIA databases were used to analyze the expression pattern of MPS-1 in HCC. Immunohistochemical staining was performed on tissue microarrays containing 169 HCC tissue samples to examine the expression of MPS-1. In addition, univariate and multivariate Cox regression analyses and Kaplan–Meier analysis were used to verify the correlation between clinicopathological factors in HCC patients and its clinical prognostic significance.
Results: MPS-1 was more highly expressed in HCC than in normal tissues, and MPS-1 expression was correlated with serum AFP levels (P = 0.003), liver cirrhosis (P = 0.024), tumor embolus (P = 0.009) and tumor recurrence (P < 0.003). MPS-1 was an independent prognostic factor for the overall survival of HCC (HR, 1.92; 95% CI, 1.01– 3.68), and a higher expression of MPS-1 predicted poorer survival. Furthermore, high expression of MPS-1 indicated a poor prognosis in patients with AFP positivity, cirrhosis or HBsAg positivity.
Conclusion: These findings demonstrate that MPS-1 is highly expressed in HCC and serves as an independent prognostic marker, highlighting the potential role of MPS-1 as a novel biomarker and therapeutic target for HCC.
Keywords: hepatocellular carcinoma, ribosomal protein, MPS-1, biomarker, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common malignancy and third leading cause of cancer-related death among all malignant tumors in China.1 Unfortunately, most cases are diagnosed at advanced stages and are not suitable for curative surgery. Even after curative treatment, the long-term prognosis remains unsatisfactory due to the high recurrence rate.2 Therefore, it is important to find valuable biomarkers for predicting the prognosis of HCC.
Ribosomes are biological nanomachines that synthesize all proteins within a cell.3 As an important component of ribosomes, ribosomal proteins can synthesize proteins and play a crucial role in cell cycle checkpoints, cell apoptosis and organismal development.4,5 Recent data have shown that they have extra-ribosomal functions apart from ribosome biogenesis and protein biosynthesis.6 Furthermore, the expression of ribosomal proteins is correlated with the development, progression and metastasis of tumors. Xie et al reported that overexpression of ribosomal protein LP1 was associated with cellular proliferation in HCC cells.7 Guo et al indicated that ribosomal protein S15a induced angiogenesis and promoted HCC through the Wnt/beta-catenin pathway.8
Metallopanstimulin-1 (MPS-1), also known as RPS27 belonging to the ribosomal protein S27E family, is a multifunctional ribosomal protein that is composed of 84 amino acids.9 MPS-1 is closely related to the development and progression of malignant tumors, such as breast cancer, gastric carcinoma, and head and neck tumors,10–14 and is involved in many biological processes, such as transcriptional regulation and DNA damage repair. MPS-1 protein contains one “zinc finger” domain of the C4 type, which could specifically bind to the cAMP response element and regulate their expression.15 MPS-1 was found to be elevated in many types of tumors, and it is frequently studied as a disease biomarker or a therapeutic molecular target. Feldheim et al revealed that MPS-1 was overexpressed in WHO grade II/III glioma and in GBM compared to NB at the mRNA and protein levels.16 Ganger et al found that MPS-1 was involved in both progression toward malignancy in regenerating cirrhotic nodules and HCC development, whereas the underlying molecular mechanism was unclear. However, the study was limited by the small number of patients and the use of nonspecific antibodies.17 Another study found that MPS-1 was elevated to high levels in 5 out of 6 primary HCC patients.11 However, to date, few studies have focused on the role of MPS-1 in HCC and its relationship with the prognosis of HCC.
In this study, we investigated the expression of MPS-1 in HCC and normal tissues through an existing database. Furthermore, we confirmed the expression pattern of MPS-1 in 169 HCC samples by immunohistochemical staining (IHC), which displayed a correlation between clinical variables and the expression of MPS-1 and demonstrated the clinical prognostic significance of MPS-1. This study, for the first time, provided theoretical support for further studies on the function and mechanism of MPS-1 in the development and progression of HCC.
Materials and Methods
Expression of MPS-1 in Oncomine, GEO and GEPIA Databases
Oncomine is a cancer microarray database and integrated data-mining platform containing 715 datasets and more than 9000 cancer and normal samples, which can help to analyze differentially expressed genes, predict coexpressed genes and identify novel biomarkers and therapeutic targets of tumors.18 In this study, Oncomine was utilized to screen the expression of MPS-1 in HCC datasets, and the search conditions were set as follows: 1) Gene: MPS-1 (RPS27); 2) Analysis type: Differential expression: Cancer vs Normal analysis; 3) Cancer type: Liver cancer; 4) Datasets: ORDER BY Overexpression; 5) Threshold by: P<1E-4, fold change ≥ 2. TCGA database:TCGA-LIHC (Cancer Genome Atlas -Liver hepatocellular carcinoma). GEO (Gene Expression Omnibus) database:GSE7629 and GSE84598.
GEPIA (Gene Expression Profiling Interactive Analysis) is an interactive platform that provides functions, including differential expression analysis, profiling plotting and correlation analysis based on the RNA-Sequencing data of 9736 tumors and 8587 normal samples from The Cancer Genome Atlas (TCGA) and GTEx datasets.19 The search conditions were set as follows: 1) Gene name: MPS-1; 2) Dataset selection: LIHC; 3) Express DIY: Stage plot; and 4) Matched Normal data: Match TCGA normal and GTEx data.
HCC Tissue Samples
Tissue microarrays contained 169 hepatic cancer tissue samples from HCC patients who underwent surgery at hospitals that cooperated with Shanghai Outdo Biotech between 2006 and 2008. The 169 samples were obtained from 141 men and 28 women, ranging from 27 to 80 years of age (median: 53 years) with 1–83 months of follow-up information. This study was approved by the institutional Review Board of Zhongshan Hospital, Fudan University. All human participants provided informed consent.
Immunohistochemical Staining
Immunohistochemical (IHC) staining for MPS-1 was performed on HCC tissue microarrays. After deparaffinization and antigen retrieval, slides were blocked with 5% bovine serum albumin and incubated with antibodies against MPS-1 (Abcam, Cambridge, UK; ab4385) at 4 °C overnight. After washing with PBS, a biotinylated secondary antibody (1:200) was added, incubated at 37 °C for 30 min, and visualized by the standard avidin-biotinylated peroxidase complex method. The expression of MPS-1 was examined by light microscopy, and five random IHC images of each slide were captured. The percentage of positive tumor cells and the intensity of immunostaining were quantified and graded according to the following criteria: 0, less than 5% positive cells; 1, 5–24% positive cells; 2, 25–49% positive cells; 3, 50–74% positive cells; and 4, ≥75% positive cells. The intensity of staining for MPS-1 was scored as 1 for yellow staining, 2 for claybank and 3 for sepia staining.10 The percentage of positive tumor cells and staining intensity were multiplied to produce a weighted score to represent the expression of each case. The cases with weighted scores of 0–5 were termed negative or weakly positive, 6–8 as moderately positive and 9–12 as intensively positive.20
Statistical Analysis
Student’s t-test was performed when comparing two groups. The correlations of the clinical factors of the hepatic cancer patients were analyzed using the chi-squared test. The survival data were used to draw Kaplan–Meier curves, and the differences among the groups were analyzed by a log-rank assay. Cox proportional hazards modeling was applied to assess the prognostic significance of HCC patients. SPSS statistics 26.0 were used for the analyses. P < 0.05 was considered statistically significant.
Results
MPS-1 is Highly Expressed in HCC
When searching the expression of MPS-1-related studies from the Oncomine database, 7 HCC datasets were found to meet the search conditions, and 1127 samples were involved.18 Three datasets with larger sample sizes and representative characteristics were selected: Wurmbach,21 Guichard liver,22 and Rosser liver.23 When analyzing these datasets, we found that the expression of MPS-1 was enhanced in HCC tissues compared with normal tissues (Figure 1A). Furthermore, the analyzed results in the TCGA and GEO databases were in accordance with the above analyses (Figure 1B).
Expression of MPS-1 in HCC Tissues
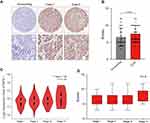
To further illustrate the expression pattern of MPS-1, we performed IHC for MPS-1 using tissue microarrays that contained 169 HCC tissue samples. The HCC tissues displayed a relatively higher expression of MPS-1 than the normal tissues (Figure 2A and B). Additionally, to observe whether the expression of MPS-1 was positively associated with pathological stage, we used the GEPIA database to analyze the correlations. The results showed that the expression of MPS-1 was not elevated with increasing stage (Figure 2C). To confirm the results, we analyzed the expression of MPS-1 in each stage of HCC tissue samples, and the results were in accordance with the above conclusions (Figure 2D). This may be because MPS-1 expression is relatively high in low-stage HCC tissues, and high-stage HCC does not enhance its expression.
The clinical and pathologic characteristics of 169 hepatic cancer patients are listed in Table 1. The results clearly showed a positive correlation of MPS-1 intensity with the serum AFP level, cirrhosis status, cancer embolus and recurrence. However, there were no correlations between MPS-1 intensity and other clinical variables, such as sex, age, HBsAg and tumor stage.
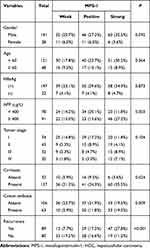 |
Table 1 Expression of MPS-1 in HCC and Its Correlation with Clinical Variables |
Relationship Between the Expression of MPS-1 and Prognosis of HCC Patients
Because MPS-1 is highly expressed in HCC, we speculated that the high expression of MPS-1 predicted poor survival in HCC patients. To test this speculation, the indicated 169 patients were analyzed for overall survival (OS) and progression-free survival (PFS). HCC patients with high MPS-1 expression had significantly poorer OS and PFS than those with low MPS-1 expression (Figure 3). Furthermore, we found that high expression of MPS-1 indicated a poor prognosis in patients with either low or high levels of serum AFP (Figure 4A). Patients with higher MPS-1 expression had a worse prognosis but not patients without cirrhosis (Figure 4B). Similarly, in patients with positive HBsAg, the high expression of MPS-1 indicated a detrimental prognosis compared with patients with low expression of MPS-1 (Figure 4C).
Further univariate and multivariate Cox analyses showed that high expression of MPS-1 was an independent prognostic factor for poor survival in HCC patients (HR 1.92, 95% CI 1.01–3.68, P=0.035; Table 2). These results suggest that the expression of MPS-1 is an important factor that affects the prognosis of HCC patients.
 |
Table 2 Univariate and Multivariate Cox Regression Analysis of Different Prognosis Factors in HCC Patients |
Discussion
MPS-1 is reported to be overexpressed in various kinds of cancers and may be involved in tumorigenesis. However, little is known about its role in HCC. In the current study, we provide strong evidence for the first time that MPS-1 is overexpressed in HCC tissues and correlates with poor prognosis of HCC patients.
To explore the expression pattern of MPS-1 in HCC, we first adopted bioinformatics analysis by using datasets from the Oncomine and GEIPIA databases, which demonstrated that MPS-1 was highly expressed in HCC tissues compared with normal tissues, and the expression had no relationship with pathological stages. In line with the database analysis, IHC staining of tissue microarrays containing 169 HCC tissues showed that MPS-1 is overexpressed in cancer tissues. We showed that the expression of MPS-1 correlated with the serum AFP level, cirrhosis status, cancer embolus and tumor recurrence. AFP level, cancer embolus and tumor recurrence were indicators for the aggressive biological behavior of HCC,24 which agrees with the expression pattern of MPS-1. MPS-1 was reported to be intensively expressed in regenerating cirrhotic nodules and poorly differentiated HCC and involved in the progression toward malignancy.17 The NF-κB and epithelial mesenchymal transition pathways may contribute to the mechanism of this progress;10,25 however, the exact mechanism has to be further explored.
In clinical practice, no studies have examined the relationship between MPS-1 expression and the prognosis of HCC patients. Here, we showed that OS and PFS were poor in patients with high MPS-1 expression. Interestingly, as MPS-1 expression was correlated with serum AFP levels, regardless of serum AFP at a low or high level, the prognosis of HCC patients with high expression of MPS-1 was poor. In patients with cirrhosis, patients with higher MPS-1 expression displayed poorer prognosis. This partly indicated that MPS-1 expression significantly affected the survival of HCC patients.
There are few reports about the role of MPS-1 in hepatocarcinogenesis. In chronic hepatitis, hepatocytes had very weak MPS-1 immunostaining; however, the staining for MPS-1 was strong in hepatocytes in patients with cirrhosis. In well-differentiated HCC, MPS-1 presence was intense at the periphery of the malignant nodule. In poorly differentiated HCC, MPS-1 expression was intense in malignant hepatocytes invading septal tissues. These results suggest that MPS-1 may be involved in both progression toward malignancy in regenerating cirrhotic nodules and in subsequent steps of hepatocarcinogenesis.17 However, the mechanisms underlying this process are unknown. In gastric cancer, we found that knockdown of MPS-1 was related with reduced invasion and migration; in contrast, overexpression of MPS-1 enhanced the invasion and migration of gastric mucosal epithelial cells through regulation of integrin beta 4.10 Furthermore, MPS-1 could be used as an indicator of prognosis in gastric cancer patients, as high expression of MPS-1 is associated with poor prognosis. Additionally, inhibition of MPS-1 induced apoptosis of gastric cancer cells through the MPS-1/NF-κB/Gadd45β pathway, which may serve as a therapeutic target in the treatment of gastric cancer.25 However, whether these pathways contribute to the carcinogenesis of HCC with high MPS-1 expression is still unknown and needs further study.
Conclusions
Our work identified that MPS-1 is highly expressed in patients with HCC, especially in patients with higher serum AFP level, cirrhosis, cancer embolus and recurrence. Importantly, high expression of MPS-1 indicated poor prognosis and was an independent prognostic factor for the overall survival of HCC patients. These results demonstrated the potential role of MPS-1 as a novel biomarker for the prognosis of HCC and a therapeutic target for HCC in the future.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr. 2020;9(4):452–463. doi:10.21037/hbsn-20-480
2. Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11(4):317–370.
3. Dedkova LM, Hecht SM. Expanding the scope of protein synthesis using modified ribosomes. J Am Chem Soc. 2019;141(16):6430–6447. doi:10.1021/jacs.9b02109
4. Huang G, Li H, Zhang H. Abnormal expression of mitochondrial ribosomal proteins and their encoding genes with cell apoptosis and diseases. Int J Mol Sci. 2020;21(22):8879. doi:10.3390/ijms21228879
5. Petibon C, Malik Ghulam M, Catala M, Abou Elela S. Regulation of ribosomal protein genes: an ordered anarchy. Wiley Interdiscip Rev RNA. 2020;12:e1632.
6. Catez F, Dalla Venezia N, Marcel V, et al. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem Pharmacol. 2019;159:74–81. doi:10.1016/j.bcp.2018.11.014
7. Xie C, Cao K, Peng D, Qin L. RPLP1 is highly expressed in hepatocellular carcinoma tissues and promotes proliferation, invasion and migration of human hepatocellular carcinoma Hep3b cells. Exp Ther Med. 2021;22(1):752. doi:10.3892/etm.2021.10184
8. Guo P, Wang Y, Dai C, et al. Ribosomal protein S15a promotes tumor angiogenesis via enhancing Wnt/beta-catenin-induced FGF18 expression in hepatocellular carcinoma. Oncogene. 2018;37(9):1220–1236. doi:10.1038/s41388-017-0017-y
9. Fernandez-Pol JA, Klos DJ, Hamilton PD. A growth factor-inducible gene encodes a novel nuclear protein with zinc finger structure. J Biol Chem. 1993;268(28):21198–21204. doi:10.1016/S0021-9258(19)36910-8
10. Yang ZY, Jiang H, Qu Y, et al. Metallopanstimulin-1 regulates invasion and migration of gastric cancer cells partially through integrin beta4. Carcinogenesis. 2013;34(12):2851–2860. doi:10.1093/carcin/bgt226
11. Fernandez-Pol JA. Increased serum level of RPMPS-1/S27 protein in patients with various types of cancer is useful for the early detection, prevention and therapy. Cancer Genomics Proteomics. 2012;9(4):203–256.
12. Stack BC, Hollenbeak CS, Lee CM, et al. Metallopanstimulin as a marker for head and neck cancer. World J Surg Oncol. 2004;2:45. doi:10.1186/1477-7819-2-45
13. Ganger DR, Hamilton PD, Fletcher JW, Fernandez-Pol JA. Metallopanstimulin is overexpressed in a patient with colonic carcinoma. Anticancer Res. 1997;17(3C):1993–1999.
14. Sundblad AS, Ricci L, Xynos FP, Fernandez-Pol JA. Metallopanstimulin/S27 ribosomal antigen expression in stages I and II breast cancer: its relationship with clinicopathologic factors. Cancer Genomics Proteomics. 2005;2(1):53–60.
15. Liu Y, Ma J, Zhang L, Lin J, Liu X. Overexpressed MPS-1 contributes to endometrioma development through the NF-kappaB signaling pathway. Reprod Biol Endocrinol. 2021;19(1):111. doi:10.1186/s12958-021-00796-z
16. Feldheim J, Kessler AF, Schmitt D, et al. Ribosomal protein S27/metallopanstimulin-1 (RPS27) in glioma-A new disease biomarker? Cancers (Basel). 2020;12(5):1085. doi:10.3390/cancers12051085
17. Ganger DR, Hamilton PD, Klos DJ, et al. Differential expression of metallopanstimulin/S27 ribosomal protein in hepatic regeneration and neoplasia. Cancer Detect Prev. 2001;25(3):231–236.
18. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112
19. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi:10.1093/nar/gkz430
20. Yang Z, Li C, Yan C, et al. KIF14 promotes tumor progression and metastasis and is an independent predictor of poor prognosis in human gastric cancer. Biochim Biophys Acta Mol Basis Dis. 2019;1865(1):181–192. doi:10.1016/j.bbadis.2018.10.039
21. Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi:10.1002/hep.21622
22. Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi:10.1038/ng.2256
23. Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi:10.1158/0008-5472.CAN-10-2607
24. Zhan YT, Li L, Zeng TT, et al. SNRPB-mediated RNA splicing drives tumor cell proliferation and stemness in hepatocellular carcinoma. Aging (Albany NY). 2020;13(1):537–554. doi:10.18632/aging.202164
25. Yang ZY, Qu Y, Zhang Q, et al. Knockdown of metallopanstimulin-1 inhibits NF-kappaB signaling at different levels: the role of apoptosis induction of gastric cancer cells. Int J Cancer. 2012;130(12):2761–2770. doi:10.1002/ijc.26331
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.