Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Exploring the Mechanisms and Molecular Targets of Taohong Siwu Decoction for the Treatment of Androgenetic Alopecia Based on Network Analysis and Molecular Docking
Authors Tan X, He Y, Ou Y, Xiong X , Deng Y
Received 4 March 2022
Accepted for publication 22 June 2022
Published 1 July 2022 Volume 2022:15 Pages 1225—1236
DOI https://doi.org/10.2147/CCID.S361820
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Xiaoqi Tan,1,* Yuxin He,1,* Yongliang Ou,2 Xia Xiong,1 Yongqiong Deng1
1Department of Dermatology STD, the Affiliated Hospital of Southwest Medical University, Luzhou, People’s Republic of China; 2Health Management Center, Luzhou People’s Hospital, Luzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongqiong Deng; Xia Xiong, Department of Dermatology STD, the Affiliated Hospital of Southwest Medical University, No. 25, Taiping Street, Luzhou, 646000, Sichuan Province, People’s Republic of China, Email [email protected]; [email protected]
Purpose: Taohong Siwu decoction (THSWD) is traditionally used to treat androgenic alopecia (AGA) in clinical practice of traditional Chinese medicine. This study used a network pharmacology approach to elucidate the molecular mechanism governing the effect of THSWD on AGA.
Materials and Methods: The major active components and their corresponding targets of THSWD were screened. AGA-related targets were obtained by analyzing the differentially expressed genes between AGA patients and healthy individuals. The protein–protein interaction networks of putative targets of THSWD and AGA-related targets were visualized and merged to identify the candidate targets for THSWD against AGA. Gene ontology (GO) biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for core targets were performed. Finally, the key effective components and core targets screened were verified by molecular docking.
Results: In this study, 69 compounds and 202 compound targets of THSWD, as well as 1158 disease targets, were screened. Forty-five interactive targets were identified for constructing the “ingredient-targets” network. The functional annotations of target genes were found to be related to oxidative stress, reactive oxygen species, and hydrogen peroxide. Pathways involved in the treatment of AGA included apoptosis and PI3K-AKT signaling pathways. The luteolin, quercetin, kaempferol, baicalein, and beta-carotene were identified as the vital active compounds, and AKT1, TP53, JUN, CASP3 and MYC were considered as the core targets. Assessment of molecular docking revealed that these active compounds and targets had good-binding interactions.
Conclusion: The results indicated that the effects of THSWD against AGA may be related to anti-inflammation and anti-oxidation properties of the compounds through the specific biological processes and the related pathways.
Keywords: Taohong Siwu decoction, network pharmacology, mechanism prediction, androgenetic alopecia
Introduction
Androgenetic alopecia (AGA) is the most diagnosed hair loss dysfunction, which was characterized by progressive miniaturization of the hair follicle leading to vellus transformation of terminal hair.1 In China alone, hair loss affects more than 100 million men.2 Although AGA is not considered a life threatening disease but it can cause psycho-emotional disturbances and a significant impact on the quality of life of many patients.3 Although the hair loss in AGA are generally thought to be driven by genetic factors and androgen, the underlying molecular pathophysiology remains unclear, thus limiting effective treatments.4 For the past few years, there were only two drugs, finasteride and minoxidil, approved by the U.S. Food and Drug Administration (FDA) for effectively management of AGA.5 Topical minoxidil shortens telogen, causing premature entry of resting hair follicles into the anagen phase. Finasteride is a 5α-reductase type II inhibitor, it prevents the conversion of testosterone to dihydrotestosterone (DHT). However, once the medication is discontinued, hair loss persists and the exact mechanism of minoxidil is unclear. Additionally, the sexual dysfunction including decreased libido, decreased ejaculation volume, and ejaculation disorder caused by finasteride and the skin irritation, dizziness, and tachycardia due to minoxidil has been reported in many patients taking these conventional drugs.6,7
Currently, traditional Chinese medicine (TCM) was recommended as a potential effective auxiliary strategy to treat chronic diseases including AGA. You et al8 has conducted a meta-analysis including 30 Randomized controlled trials (RCTs) involving 2615 patients to evaluate the curative efficacy and safety of TCM for treating AGA and found that the total efficacy was significantly higher, the total symptom score markedly reduced in the TCM group, when compared with that in the conventional medicine group. In addition, no significant differences were observed between the two groups in terms of adverse events.8,9 In fact, TCM therapies have been widely used for the treatment of AGA for two thousand years and could date back to the Qin Dynasty. After thousands of years of development, ancient Chinese medicine has accumulated a large number of effective traditional Chinese herbal formulas and rich experience in AGA treatment. It has been shown that functional deficiency of liver and kidney, deficiency of qi and blood, qi stagnation and blood stasis, and blood-heat are the main patterns linked to alopecia, based on TCM pattern identification.10–12
Taohong Siwu decoction (THSWD), an improved formula of Siwu decoction, is a mixture of 6 Chinese medicine extracts including Persicae Semen (Taoren, TR, the dried ripe seed of Prunus persica (L.), Batsch or Prunus davidiana (Carr.) Franch.), Carthami Flos (Honghua, HH, the dried Carthamus tinctorius L.), Angelica sinensis radix (Danggui, DG, the dried root of Angelica sinensis), Chuanxiong Rhizoma (Chuanxiong, CX, the dried rhizome of Ligusticum chuanxiong Hort.), Paeoniae Radix Alba (Baishao, BS, the dried root of Paeonia lactiflora Pall.), Rehmanniae Radix Praeparata (Shudihuang, SDH, the dried root of Rehmannia glutinosa Libosch.). In TCM, THSWD is frequently used to treat the deficiency and stasis of qi and blood as well as the functional deficiency of liver and kidney, which were the main pathogenesis of AGA. Recently, association rule mining and network analysis have been performed to analyze the combinations of medicinal herbs used to treat alopecia and have identified DG, CX, BS, SDH, the primary ingredients of THSWD, as the most frequently used herbs.12 However, the pharmacological mechanisms and molecular targets of THSWD for the treatment of AGA are not clear, which is the main factor restricting its wider use.
In the present study, network pharmacology and molecular docking were conducted to explore the mechanism of THSWD in treating AGA (Figure 1). Firstly, the active compounds of THSWD and their targets were searched and screened. Subsequently, the AGA-related signaling pathways were obtained by analyzing the differentially expressed genes between AGA patients and healthy individuals. The mechanisms of action underlying THSWD for the treatment of AGA were analyzed by GO and pathway analysis. Finally, the key effective components and targets screened were verified by molecular docking, and the complex network of THSWD in treating AGA was comprehensively discussed.
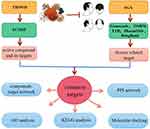 |
Figure 1 The whole framework of this study based on network pharmacology and molecular docking for investigating pharmacological mechanisms of THSWD acting on AGA. |
Methods and Materials
Active Ingredients and Potential Targets Screening
We identified the chemical composition of the six herbs (TR, HH, BS, DG, CX, SDH) in THSWD from the Traditional Chinese Medicine Database and Analysis Platform (TCMSP, https://tcmsp-e.com/),13 by establishing preset criteria of oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18 in absorption, distribution, metabolism and excretion (ADME) to screen for the active ingredients. Then, the candidate compounds were imported into the TCMSP platform to identify the corresponding targets of THSWD. With the help of the protein database UniProt, (https://www.uniprot.org/), we converted the related targets into the gene names.14 Target information was set as reviewed, and removed repetitive and non-human targets. Afterwards, the target gene-set of THSWD was acquired.
Identification of AGA-Related Targets
The differentially expressed genes of AGA patients were obtained from 5 databases as follows: GeneCards (https://www.genecards.org/), OMIM (https://omim.org/), DrugBank (https://go.drugbank.com/), PharmGKB (https://www.pharmgkb.org/), and TTD (http://db.idrblab.net/ttd/). The significance threshold of P value was set to <0.05, and the |log fold change| was set to > 1 to obtain the differentially expressed genes, considered as the AGA-related targets. In addition, we took a combination of the search results to construct an AGA relevant gene database.
Protein–Protein Interaction Network Construction
The potential targets of THSWD for the treatment of AGA were obtained by intersecting the THSWD target gene set and the AGA-related target gene database. The candidate targets were obtained by a Venn diagram and introduced into the STRING database (https://string-db.org/) to construct protein–protein interaction (PPI) network.15 We selected multiple proteins and set the parameter as moderate confidence (0.400). The organism was limited to “Homo sapiens”, and the “string_interactions.tsv” file was kept.
Construction of a Compound-Target Network and Critical Sub-Network
The compound-target network was constructed to clarify the relationship between active compounds and potential targets. The “string_interactions. tsv” was imported into Cytoscape 3.8.0 software for visualization analysis. The Network-Analyzer setting from Cytoscape 3.8.0 software was applied for analyzing the topological parameters,16 while the core targets were collected according to the Degree-value.17 Nodes in the network referred to drug chemical components or targets, and edges represented the relationship between nodes.
We applied two methods to investigate the critical sub-network. Firstly, we filtered genes with topological importance by calculating Degree Centrality (DC),18 Closeness Centrality (CC),19 Betweenness Centrality (BC),20 Eigenvector Centrality (EC), Local Average Centrality (LAC), and Network Centrality (NC). After two filters, the final core targets were selected and the critical subnetwork was established. Another approach we used to conduct critical sub-network was CytoHubba plugin. The top 10 important genes in the network are obtained through one screening.
GO and KEGG Enrichment Analysis
GO and KEGG enrichment analysis were employed to explore the potential function of the candidate targets. GO enrichment analysis includes biological process, cellular composition, and molecular function, whereas KEGG enrichment analyzes the potential biological pathways and functions associated with the targets. A variety of bioinformatic analyses and visualization of the results can be achieved using the software R project. In the programming language, the adjusted P-value <0.05 and q-value < 0.05 were set.
Molecular Docking Technology
We took an intersection of the key sub-networks obtained by two different methods to obtain the most significant genes, from which receptor proteins were selected. We downloaded the structure of core receptor proteins in PDB database (http://www.rcsb.org/pdb/home/home.do), and then PyMOL software was performed to simulate the dehydration and separation of ligands from the receptor protein. AutoDockTools was used to carry out hydrogenation. According to the degree value in the compound-target network, we used the top active ingredients as the molecular ligand. The 2D structure for the molecule ligands which was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), was converted to 3D structure by Chem3D software. Chem3D software was used to calculate and export the 3D structure by minimizing energy associated with the molecular structure. The original PBD file format was converted to the PBDQT file format recognized by the AutoDock Vina program. The receptor proteins with higher significance in the key sub-network and the corresponding molecular ligands with more value in the compound-target network were selected. After that, we used AutoDock Vina to calculate docking score, which indicated binding affinity. The lower the docking score, the more stable the bonding between a ligand and a protein.21
Results
Screening of Active THSWD and the Drug Related Targets
A total of 69 potential compounds of THSWD were obtained, 23 in TR, 13 in BS, 2 in SDH, 22 in HH, 2 in DG, and 7 in CX (Table S1). Meanwhile, drug-related targets were collected, 43 in TR, 79 in BS, 28 in SDH, 189 in HH, 45 in DG, and 29 in CX, and finally, there were 202 targets identified after removing duplications (Figure S1).
Screening the Potential Targets for THSWD Against AGA
Firstly, the AGA-related targets were screened from 5 databases. There are 393 AGA-related genes from Genecards, 19 genes from OMIM, 0 genes from TTD, 760 genes from PharmGkb and 11 genes from DrugBank (Figure 2A). A gene set with 1158 AGA-related genes was obtained after deleting overlapping genes and combining the research findings (Figure 2B). Then, the 202 compound targets were mapped to 1158 AGA target genes to obtain 45 common target genes, which are suggested to be the potential targets of THSWD in the treatment of AGA. The 45 common targets identified above were introduced into the STRING database to perform PPI network analysis, which contained 45 nodes and 380 edges (Figure 2C).
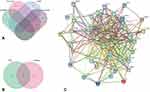 |
Figure 2 Identification of the drug–target interaction. (A) The AGA related targets from 4 databases. (B) The Venn diagram of THSWD and AGA. (C) PPI of THWSD in treating AGA. |
Building a “Compound-Target” Network
To clearly display the complex relationships between the active compounds in THSWD and AGA-related targets, a compound-target network was constructed. The network was composed of 31 active components of THSWD in the outer circle and 45 targets of AGA in the inner circle. As showed in Figure 3, we can clearly find that the compound-target network included 76 nodes and 156 edges. Luteolin (MOL000006), quercetin (MOL000098), kaempferol (MOL000422), baicalein (MOL002714), and beta-carotene (MOL002773) acted on 132, 54, 55, 35 and 18 targets, respectively. And the OB values of quercetin, luteolin, kaempferol, baicalein, and beta-carotene are 46.43%, 36.16%, 41.88%, 33.52%, and 37.18%, respectively. Therefore, the results suggest that they might be the crucial active compounds of THSWD by reason of their considerable presence in the network.
 |
Figure 3 Compound- target network of THSWD. The blue rectangular in the middle represent targets; the different color at the edge represent the compounds from different herbs, respectively. |
GO and KEGG Enrichment Analysis
GO enrichment analysis was used to clarify the function of estimated protein targets, covering the underlying biological processes, cellular components and molecular functions of the 45 target genes. According to the P-value <0.05 and q-value < 0.05, 1801 noticeably enriched GO terms were screened and the top 10 terms are shown in Figure 4A. There were 1690 biological processes, mainly involved in the response to oxidative stress, epithelial cell proliferation, the response of cells to chemical stress. Twenty-three cell components revealed membrane raft, membrane microdomain, membrane region, transcription regulator complex, etc. For 88 molecular functions, the targets were enriched in DNA-binding transcription factor binding, ubiquitin-like protein ligase binding, phosphatase binding. GO analysis showed that these target genes played an important role in oxidative stress and cell proliferation.
Furthermore, the ClusterProfiler KEGG was also used to analyze the KEGG enrichment of the 45 potential targets by R4.0.2. By setting the filter as adjusted P-value <0.05 and q-value < 0.05, we detected 138 significantly enriched KEGG terms. The KEGG signal pathway enrichment analysis was represented by a bubble diagram, and the top 30 KEGG pathways are shown in Figure 4B. These included apoptosis, lipid and atherosclerosis, human cytomegalovirus infection, Pl3K-Akt signaling pathway, AGE-RAGE signaling pathway in diabetic complications, fluid shear stress and atherosclerosis, and hepatitis B, etc. Furthermore, THSWD probably exerted the therapeutic effects on AGA by regulating signaling pathways, which included Pl3K-Akt signaling pathway, NOD-like receptor signaling pathway, IL-17 signaling pathway, AGE-RAGE signaling pathway.
The Critical Sub-Network Analysis for the Core Targets
To further identify the key targets for THSWD against AGA, 45 potential candidates were imported in CytoNCA plugin and selected by calculating DC, BC, CC and EC. A network of significant targets for THSWD against AGA was constructed and it contained 16 nodes and 110 edges (Figure 5A). The median values of DC, BC, CC, EC, LAC and NC were 14, 8.61, 0.59, 0.13, 11 and 12.50, respectively. The candidate targets were further screened and 6 core genes with DC>14, BC>1.24, CC>0.94, EC>0.25, LAC>12.14, NC>13.60 were identified. Finally, the critical sub-network consisting of six key target genes was constructed, including AKT1, TP53, JUN, CASP3, CTNNB1 and MYC (Figure 5B).
Besides, CytoHubba plugin were also employed to conduct the (the analysis of?) critical sub-network (Figure 5C). Interestingly, the top 10 key genes obtained from CytoHubba plugin overlaps with the 6 key targets screened by the CytoNCA plugin.
Molecular Docking Technology
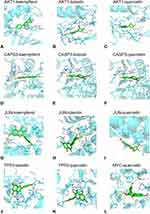
The 6 core genes (AKT1, TP53, JUN, CASP3, CTNNB1 and MYC) were chosen to conduct molecular docking analysis. After that, from the compound-target interaction network, we got homologous active compounds targeting receptor proteins. These compounds were quercetin, luteolin, kaempferol, baicalein, and beta-carotene. Molecular docking analysis was performed to further validate interactions between molecular ligands and receptor proteins. The docking score lower than −5.0 kcal/mol indicated that the conformations had good-binding interactions.22 In particular, the binding affinity of interactions found in this study were all less than −6.0 kcal/mol (Table S2). It meant that these molecular ligands could easily enter and successfully bind the active pocket of the receptor proteins with high affinity. We selected top 3 active compounds (quercetin, luteolin, kaempferol) corresponding to core targets (AKT1, TP53, JUN, CASP3 and MYC) for visual analysis of molecular docking, as shown in Figure 6.
Discussion
The drug compositions of herbs are complex in TCM, especially the more complex herbal formulations that consist of multiple herbs including numerous potential bioactive ingredients, which can interact with multiple therapeutic targets. This characteristic of TCM, the multi-component, multi-target, multi-channel and multi-gene approach, does present a tremendous challenge in exploring the potential molecular mechanism of TCM in treating diseases. Although the pharmacological properties of TCM herbs and the mechanism of their therapy on AGA have been investigated by a number of studies, the bioactive ingredients and the specific targets of its treatment of AGA still needs to be fully elucidated.8,23 Network Pharmacology is a novel approach in an attempt to solve the problems, which was firstly proposed by Li et al and employed by Liu et al to elucidate the pharmacological mechanism of Xiaoyao powder on anovulatory infertility.24,25 Network pharmacology is based on the “disease-gene-target-drug” interaction network, which could systematically and comprehensively elucidate the synergistic effects among compounds and potential mechanisms of multi-component and multiple-target drugs on human body at the molecular level, and could contribute to new strategies for TCM drug development and clinical applications.
Based on the network pharmacology approach, this study focused on the specific targets and molecular signaling pathways of THSWD acting on AGA, in which the 69 compounds, 202 compound targets and 1158 disease targets were screened to construct the compound-target-disease network. Then, the PPI networks of THSWD putative targets and AGA-related targets were structured and merged to obtain the candidate targets for THSWD against AGA. In order to find more accurate targets, 6 parameters including Degree, Betweenness Centrality, Closeness Centrality, and Local Average Connectivity were set to screen nodes and structure a new network. Forty-five common targets of compounds and disease were finally identified and used to carry out the bioinformatic analysis to elucidate the mechanisms underlying the anti-AGA effects of THSWD. The result showed the top compounds of THSWD affecting multiple targets, for example, luteolin, quercetin, kaempferol, baicalein, and beta-carotene acted on 132, 54, 55, 35 and 18 targets, respectively. Therefore, they were very likely to be the crucial pleiotropically active compounds of THSWD for treating AGA. The top three compounds, quercetin (3,5,7,3′,4′- pentahydroxyflavone), luteolin (3,4,5,7-tetrahydroxy flavone), and kaempferol are all major representatives of the flavonoid subclass of flavonols, ubiquitously present in fruits and vegetables and are the main ingredients of Honghua in THSWD. It has been reported that the three flavonoids could exert a wide range of biological effects, including antioxidant, anticarcinogenic, anti-inflammatory, anti-diabetic and antimicrobial activities.26–33 Throughout the literature, luteolin has not been reported to be used to treat AGA, but quercetin in recent study was reported to be packaged in phospholipid-polymer hybrid nanoparticles used in AGA animal models. It was shown to improve hair regrowth and inhibit hair follicles cells apoptosis, and the study suggested the role of quercetin on AGA by reducing the synthesis of prostaglandin D2, which was reported to induce miniaturization, sebaceous gland hyperplasia, and alopecia.33–36 Additionally, the hair cell proliferation induced by quercetin and kaempferol was significantly higher than that of minoxidil, supporting the development of these compounds into commercially available treatments for alopecia.37 From this study, AKT1, TP53, JUN, CASP3 and MYC were also identified as the specific targets of quercetin, luteolin and kaempferol on AGA, suggesting that the roles of the three compounds on the management of AGA could be related to their anti-inflammation and anti-oxidation properties.
The targets of THSWD against AGA were enriched in biological processes, cellular components, and molecular function by GO enrichment analysis. Results suggest that THSWD regulated some biological processes such as the cell response to oxidative stress, reactive oxygen species (ROS), and hydrogen peroxide. Oxidative stress is a result of inadequate antioxidant defense or overproduction of ROS, which has been demonstrated in promoting secretion of known hair follicle inhibitory factors in the biological process of AGA.38–42 Oxidative stress in addition accelerates the aging process as AGA deteriorates with age.41,43 So, topical or systemic antioxidants has been suggested to be the promising treatment of AGA.42 In this study, the flavonoid ingredients luteolin, quercetin, and kaempferol were identified as the crucial pleiotropically active compounds of THSWD for treating AGA, which were responsible for the strong antioxidant activities.44–47 Other active ingredients in THSWD also have been reported to have anti-inflammatory and antioxidant effects in NF-κB signaling pathway and apoptotic pathway.48
To date, the most elucidated AGA etiopathogenesis are the genetics and the androgen-mediated follicular miniaturization. However, medical treatments regarding the androgen activity modulation, such as finasteride, have sometimes shown limited efficacy in AGA.5 Probably, there are more to be understood regarding its physiopathology and potential treatments beyond androgens and anti-androgens.49 In the present study, a total of 30 KEGG pathways including apoptosis, Pl3K-Akt signaling pathway, NOD-like receptor signaling pathway, IL-17 signaling pathway, and AGE-RAGE signaling pathway were significantly enriched. Gene-pathway network was constructed to investigate the core and key target genes for THSWD against AGA. The top 6 targets of AKT1, TP53, JUN, CASP3 and MYC were suggested as the core target genes. It is known that 0p0; Caspase-3 plays an important role in apoptosis, as it is activated in the apoptotic cell by extrinsic (death ligand) and intrinsic (mitochondrial) pathways and TNF is an inflammatory cytokine that plays a well-established role and acts on several different signaling pathways through two cell surface receptors, being probably the most potent inducer of apoptosis.50 CASP3 was overexpressed in alopecia areas in patients with AGA at early stages of the disease, which indicates that apoptosis and inflammation are present in the early stages of this disorder.50 Previous studies also have shown that Akt pathways promote dermal papilla cell survival.51 Phosphorylation of Akt was lower and inflammatory factors such as IL-1β, TNF-α, IL-6 were up-regulated in the dorsal skin of AGA induced by testosterone propionate in C57BL/6 mice, which could be reversed by Radix paeoniae alba.52
In summary, this study used network pharmacological methods and techniques to explore the mechanisms of action and molecular targets of THSWD for treating AGA. THSWD may promote hair growth through the specific biological processes including responses to oxidative stress, reactive oxygen species, and hydrogen peroxide. In addition, THSWD may exert its regulatory function in the pathogenesis of AGA and the regulation of pathways including apoptosis, AGE-RAGE signaling pathway, NOD-like receptor signaling pathway and PI3K-AKT signaling pathways, which are associated with the occurrence of AGA. CASP3, MAPK1, AKT1, JUN, TNF, TP53, IL-6 and VEGF-A were the key target genes in the gene network of THSWD for the treatment of AGA. The network pharmacology appears to be a suitable approach for the study of complex TCM formulations.
Ethical Approval
The study protocol was approved by the local ethics committee of Southwest Medical University and with the 1964 Helsinki declaration.
Funding
This study was funded by the project of Southwest Medical University (granted number: NO.2021ZKMS027/2021ZKMS030) and Sichuan Medical Association (granted number: S21022).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lolli F, Pallotti F, Rossi A, et al. Androgenetic alopecia: a review. Endocrine. 2017;57(1):9–17. doi:10.1007/s12020-017-1280-y
2. Zhang Y Study of the differences on hair follicle between AGA and healthy. [PhD thesis]. Shanghai, China: Second Military Medical University; 2016.
3. Zhang M, Zhang N. Quality of life assessment in patients with alopecia areata and androgenetic alopecia in the People’s Republic of China. Patient Prefer Adherence. 2017;11:151–155. doi:10.2147/PPA.S121218
4. Martinez-Jacobo L, Villarreal-Villarreal CD, Ortiz-López R, Ocampo-Candiani J, Rojas-Martínez A. Genetic and molecular aspects of androgenetic alopecia. Indian J Dermatol Venereol Leprol. 2018;84(3):263–268. doi:10.4103/ijdvl.IJDVL_262_17
5. York K, Meah N, Bhoyrul B, Sinclair R. A review of the treatment of male pattern hair loss. Expert Opin Pharmacother. 2020;21(5):603–612. doi:10.1080/14656566.2020.172146
6. Goren A, Naccarato T. Minoxidil in the treatment of androgenetic alopecia. Dermatol Ther. 2018;31(5):e12686. doi:10.1111/dth.12686
7. Lee S, Lee YB, Choe SJ, Lee WS. Adverse sexual effects of treatment with finasteride or dutasteride for male androgenetic alopecia: a systematic review and meta-analysis. Acta Derm Venereol. 2019;99(1):12–17. doi:10.2340/00015555-3035
8. You Q, Li L, Ma X, et al. Meta-analysis on the efficacy and safety of traditional Chinese medicine as adjuvant therapy for refractory androgenetic alopecia. Evid Based Complement Calternat Med. 2019;2019:9274148. doi:10.1155/2019/9274148
9. Dhariwala MY, Ravikumar P. An overview of herbal alternatives in androgenetic alopecia. J Cosmet Dermatol. 2019;18(4):966–975. doi:10.1111/jocd.12930
10. Xuemeng DU. Study on medication and prescription compatibility law of alopecia disease (in Chinese). Nanjing University of Chin Med. 2016;19(11):2461–2475.
11. Fu-chun SI, Xian-pei MENG. Analysis of the TCM syndrome and prescription rules of alopecia and leukotrichia. CJTCMP. 2016;2016(9):3785–3788.
12. Leem J, Jung W, Kim Y, Kim B, Kim K. Exploring the combination and modular characteristics of herbs for alopecia treatment in traditional Chinese medicine: an association rule mining and network analysis study. BMC Complement Altern Med. 2018;18(1):204. doi:10.1186/s12906-018-2269-7
13. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi:10.1186/1758-2946-6-13
14. Chen Z, Lin T, Liao X, et al. Network pharmacology based research into the effect and mechanism of yinchenhao decoction against cholangiocarcinoma. Chin Med. 2021;16(1):13. doi:10.1186/s13020-021-00423-4
15. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
16. Li R, Li Y, Liang X, Yang L, Su M, Lai KP. Network pharmacology and bioinformatics analyses identify intersection genes of niacin and COVID-19 as potential therapeutic targets. Brief Bioinform. 2021;22(2):1279–1290. doi:10.1093/bib/bbaa300
17. Xiao H, Qin X, Wan J, Li R. Pharmacological targets and the biological mechanisms of formononetin for alzheimer’s disease: a network analysis. Med Sci Monit. 2019;25:4273–4277. doi:10.12659/MSM.916662
18. Missiuro PV, Liu K, Zou L, et al. Information flow analysis of interactome networks. PLoS Comput Biol. 2009;5(4):e1000350. doi:10.1371/journal.pcbi.1000350
19. Raman K, Damaraju N, Joshi GK. The organisational structure of protein networks: revisiting the centrality-lethality hypothesis. Syst Synth Biol. 2014;8(1):73–81. doi:10.1007/s11693-013-9123-5
20. Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi:10.1016/j.biosystems.2014.11.005
21. Xu L, Zhang J, Wang Y, Zhang Z, Wang F, Tang X. Uncovering the mechanism of Ge-Gen-Qin-Lian decoction for treating ulcerative colitis based on network pharmacology and molecular docking verification. Biosci Rep. 2021;41(2):BSR20203565. doi:10.1042/BSR20203565
22. Li X, Xu X, Wang J, et al. A system-level investigation into the mechanisms of Chinese traditional medicine: compound Danshen formula for cardiovascular disease treatment. PLoS One. 2012;7(9):e43918. doi:10.1371/journal.pone.0043918
23. Cho EC, Kim K. A comprehensive review of biochemical factors in herbs and their constituent compounds in experimental studies on alopecia. J Ethnopharmacol. 2020;258:112907. doi:10.1016/j.jep.2020.112907
24. Li S, Zhang ZQ, Wu LJ, Zhang XG, Li YD, Wang YY. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst Biol. 2007;1(1):51–60. doi:10.1049/iet-syb:
25. Liu H, Zeng L, Yang K, Zhang G. A network pharmacology approach to explore the pharmacological mechanism of xiaoyao powder on anovulatory infertility. Evid Based Complement Alternat Med. 2016;2016:2960372. doi:10.1155/2016/2960372
26. Jafarinia M, Sadat Hosseini M, Kasiri N, et al. Quercetin with the potential effect on allergic diseases. Allergy Asthma Clin Immunol. 2020;16:36. doi:10.1186/s13223-020-00434-0
27. Xu D, Hu MJ, Wang YQ, Cui YL. Antioxidant activities of quercetin and its complexes for medicinal application. Molecules. 2019;24(6):1123. doi:10.3390/molecules24061123
28. Reyes-Farias M, Carrasco-Pozo C. The anti-cancer effect of quercetin: molecular implications in cancer metabolism. Int J Mol Sci. 2019;20(13):3177. doi:10.3390/ijms20133177
29. Dabeek WM, Marra MV. Dietary quercetin and kaempferol: bioavailability and potential cardiovascular-related bioactivity in humans. Nutrients. 2019;11(10):2288. doi:10.3390/nu11102288
30. Eid HM, Haddad PS. The antidiabetic potential of quercetin: underlying mechanisms. Curr Med Chem. 2017;24(4):355–364. doi:10.2174/0929867323666160909153707
31. Gendrisch F, Esser PR, Schempp CM, Wölfle U. Luteolin as a modulator of skin aging and inflammation. Biofactors. 2021;47(2):170–180. doi:10.1002/biof.1699
32. Imran M, Rauf A, Abu-Izneid T, et al. Luteolin, a flavonoid, as an anticancer agent: a review [published correction appears in biomed pharmacother]. Biomed Pharmacother. 2019;112:108612. doi:10.1016/j.biopha.2019.108612
33. Ren J, Lu Y, Qian Y, Chen B, Wu T, Ji G. Recent progress regarding kaempferol for the treatment of various diseases. Exp Ther Med. 2019;18(4):2759–2776. doi:10.3892/etm.2019.7886
34. Das L, Kaurav M, Pandey RS. Phospholipid-polymer hybrid nanoparticle-mediated transfollicular delivery of quercetin: prospective implement for the treatment of androgenic alopecia. Dev Ind Pharm. 2019;45(10):1654–1663. doi:10.1080/03639045.2019.1652635
35. Garza LA, Liu Y, Yang Z, et al. Prostaglandin D2 inhibits hair growth and is elevated in bald scalp of men with androgenetic alopecia. Sci Transl Med. 2012;4(126):126ra34. doi:10.1126/scitranslmed.3003122
36. Trüeb RM. Oxidative stress in ageing of hair. Int J Trichol. 2009;1(1):6–14. doi:10.4103/0974-7753.51923
37. Taira N, Nguyen BC, Tawata S. Hair growth promoting and anticancer effects of p21-activated kinase 1 (PAK1) inhibitors isolated from different parts of Alpinia zerumbet. Molecules. 2017;22(1):132. doi:10.3390/molecules22010132
38. Cwynar A, Olszewska-Słonina D, Czajkowski R, et al. Evaluation of selected parameters of oxidative stress in patients with alopecia areata. Postepy Dermatol Alergol. 2019;36(1):115–116. doi:10.5114/pdia.2017.71237
39. Balık AR, Balık ZB, Aktaş A, Neşelioğlu S, Karabulut E, Karabulut AB. Examination of androgenetic alopecia with serum biomarkers. J Cosmet Dermatol. 2021;20(6):1855–1859. doi:10.1111/jocd.13732
40. Prie BE, Iosif L, Tivig I, Stoian I, Giurcaneanu C. Oxidative stress in androgenetic alopecia. J Med Life. 2016;9(1):79–83.
41. Upton JH, Hannen RF, Bahta AW, Farjo N, Farjo B, Philpott MP. Oxidative stress-associated senescence in dermal papilla cells of men with androgenetic alopecia. J Invest Dermatol. 2015;135(5):1244–1252. doi:10.1038/jid.2015.28
42. Kaya Erdogan H, Bulur I, Kocaturk E, Yildiz B, Saracoglu ZN, Alatas O. The role of oxidative stress in early-onset androgenetic alopecia. J Cosmet Dermatol. 2017;16(4):527–530. doi:10.1111/jocd.12300
43. Stefanadi EC, Dimitrakakis G, Antoniou CK, et al. Metabolic syndrome and the skin: a more than superficial association. Reviewing the association between skin diseases and metabolic syndrome and a clinical decision algorithm for high risk patients. Diabetol Metab Syndr. 2018;10:9. doi:10.1186/s13098-018-0311-z
44. Chen Q, Wang D, Tan C, Hu Y, Sundararajan B, Zhou Z. Profiling of flavonoid and antioxidant activity of fruit tissues from 27 Chinese local citrus cultivars. Plants. 2020;9(2):196. doi:10.3390/plants9020196
45. Anand David AV, Arulmoli R, Parasuraman S. Overviews of biological importance of quercetin: a bioactive flavonoid. Pharmacogn Rev. 2016;10(20):84–89. doi:10.4103/0973-7847.194044
46. Wang J, Fang X, Ge L, et al. Antitumor, antioxidant and anti-inflammatory activities of kaempferol and its corresponding glycosides and the enzymatic preparation of kaempferol. PLoS One. 2018;13(5):e0197563. doi:10.1371/journal.pone.0197563
47. Ahmadi SM, Farhoosh R, Sharif A, Rezaie M. Structure-antioxidant activity relationships of luteolin and catechin. J Food Sci. 2020;85(2):298–305. doi:10.1111/1750-3841.14994
48. Duan X, Pan L, Peng D, et al. Analysis of the active components and metabolites of Taohong Siwu decoction by using ultra high performance liquid chromatography quadrupole time-of-flight mass spectrometry. J Sep Sci. 2020;43(22):4131–4147. doi:10.1002/jssc.202000498
49. Katzer T, Leite Junior A, Beck R, da Silva C. Physiopathology and current treatments of androgenetic alopecia: going beyond androgens and anti-androgens. Dermatol Ther. 2019;32(5):e13059. doi:10.1111/dth.13059
50. Martinez-Jacobo L, Ancer-Arellano CI, Ortiz-Lopez R, et al. Evaluation of the expression of genes associated with inflammation and apoptosis in androgenetic alopecia byTargeted RNA-Seq. Skin Appendage Disord. 2018;4(4):268–273. doi:10.1159/000484530
51. Stamatas GN, Wu J, Pappas A, et al. An analysis of gene expression data involving examination of signaling pathways activation reveals new insights into the mechanism of action of minoxidil topical foam in men with androgenetic alopecia. Cell Cycle. 2017;16(17):1578–1584. doi:10.1080/15384101.2017.1327492
52. Zhang T, Cao S, Yuan H, Park S. Alleviation of androgenetic alopecia with aqueous Paeonia lactiflora and poria cocos extract intake through suppressing the steroid hormone and inflammatory pathway. Pharmaceuticals. 2021;14(11):1128. doi:10.3390/ph14111128
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.