Back to Journals » International Journal of General Medicine » Volume 15
Exploration in the Therapeutic and Multi-Target Mechanism of Ketamine on Cerebral Ischemia Based on Network Pharmacology and Molecular Docking
Authors Xiong L , Liu SC, Huo SY, Pu LQ, Li JJ, Bai WY, Yang Y, Shao JL
Received 6 January 2022
Accepted for publication 12 April 2022
Published 20 April 2022 Volume 2022:15 Pages 4195—4208
DOI https://doi.org/10.2147/IJGM.S345884
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Li Xiong,1,* Shi-Cheng Liu,2,* Si-Ying Huo,1 Lan-Qing Pu,1 Jun-Jie Li,1 Wen-Ya Bai,1 Yuan Yang,1 Jian-Lin Shao1
1Department of Anesthesiology, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, 650032, People’s Republic of China; 2Department of Oncology, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, 650032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jian-Lin Shao, Department of Anesthesiology, The First Affiliated Hospital of Kunming Medical University, 295 Xichang Road, Kunming, Yunnan, 650032, People’s Republic of China, Email [email protected]
Background: Ketamine is famous for its dissociative anesthetic properties. It is also analgesic, anti-inflammatory and anti-depressant, and even has a cerebral protective effect. We searched the evidence of the correlation between ketamine target and clinical efficacy and utilized network pharmacology to gather information about the multi-target mechanism of ketamine against cerebral ischemia (CI). We found that ketamine’s clinical significance may be more extensive than previously thought.
Methods: The drug target of ketamine and CI-related genes were predicted by SwissTargetPrediction, DrugBank, PubChem, GeneCards and DisGeNET databases. The intersection of ketamine’s drug-targets and CI-related genes was analyzed by using GO and KEGG. We predicted the molecular docking between the potential target and ketamine.
Results: The results indicated that the effect of ketamine on CI was primarily associated with the target of α-synuclein (SNCA), muscarinic acetylcholine receptor M1 (CHRM1) and nitric oxide synthase 1 (NOS1). It principally regulates the signal pathways of circadian transmission, calcium signaling pathway, dopaminergic synapse, cholinergic synapse and glutamatergic synapse. Molecular docking analysis exhibited that hydrogen bond and Pi-Pi interaction were the predominant modes of interaction.
Conclusion: There are protein targets affected by ketamine in the treatment of CI. Three pivotal targets involving 298 proteins, SNCA, CHRM1 and NOS1, have emerged as multi-target mechanisms for ketamine in CI therapy. Similarly, this study also provides a new idea for introducing network pharmacology into the evaluation of multi-targeted drugs for CI and cerebral protection.
Keywords: ketamine, cerebral ischemia, treatment target, network pharmacology
Introduction
Cerebral ischemia (CI) is a widespread acute cerebrovascular disease, which is the second leading cause of death in the world,1 and China has the highest burden of stroke in the world.2 Ischemic stroke is characterized by high incidence rate, high disability rate and high mortality rate.3,4 The critical aspect of treatment is to reduce or prevent the nerve injury caused by CI.5,6 The neuroprotective effects of CI have been studied for more than half a century, especially in the past 30 years.7,8 Nevertheless, the really effective method is still under exploration. Therefore, it is imperative to seek new treatment methods.
Ketamine is a classic intravenous anesthetic. Its anesthetic and analgesic properties are typically attributed to its direct inhibition of N-methyl-D-aspartate receptor (NMDAR).9 Not only that but it also has affinity for many other receptors, including γ-aminobutyric acid, dopamine, serotonin, opioid and cholinergic receptors, as well as voltage-gated sodium and hyperpolarized cyclic nucleotide gated channels. Hence, its clinical significance may be broader than previously thought.10,11 In recent years, ketamine has made significant breakthroughs as an antidepressant.12,13 In the treatment of CI, although previous studies have demonstrated that ketamine can ameliorate the behavioral and neuroprotective effects on animal models.14 It may be achieved by affecting the NF-κB signaling pathway, reducing TNF-α, IL-6 and increasing the content of IL-10.15 It may also activate mTOR signal transduction pathway through p-mTOR and down-regulate the expression of key autophagic proteins (such as LC3 and beclin-1), while attenuating secondary brain injury and reducing inflammatory response.16 Nevertheless, the precise mechanism of ketamine for CI is not known.
Along with the rapid development of bioinformatics, systems biology and pharmacology, web-based drug discovery and evaluation databases are considered to be an effective and cost-effective method for drug development.17,18 The combination of online pharmacology and bioinformatics tools has broadened the understanding of potential drug targets and their intersection with disease-specific key genes.19,20 Experimental data on gene, protein and metabolic interactions are combined with clinical knowledge of disease and pharmacology to explore new therapeutic approaches, which provides a macroscopic way to understand and treat complex diseases.21 Understanding pharmacokinetic and pharmacodynamic models will accelerate the discovery of new drugs with better therapeutic outcomes by combining them with pathway and network analysis.22,23 These approaches may mitigate the risk of single-target and symptom-based drug discovery and treatment failure in the future.24
In this paper, we applied a network pharmacology approach to integrate the multi-target mechanism of ketamine against CI, and further explore how ketamine can be effective against CI and facilitate new drug development using evidence of the relevance of ketamine targets to CI disease targets. In general, the interpretation of Ketamine‘s network pharmacology will hopefully lead to the development of new drugs suitable for the clinical treatment of CI disease.
Materials and Methods
Statement of Human Data
In this study, the databases containing human data include SwissTargetPrediction,25,26 DrugBank,27 PubChem,28 PDB29 ZINC30 GeneCards and DisGeNET.31,32 This study made use of data from public datasets and was approved by the human Experiment Ethics Committee of the First Affiliated Hospital of Kunming Medical University.
Identification of Ketamine Target Genes
In order to avoid missing all the potential target proteins of Ketamine as far as possible, we searched for “ketamine” in three databases: SwissTargetPrediction,25,26 DrugBank27 and PubChem.28 All the results were limited to “Homo sapiens”, and 39 human target genes that might be targeted by ketamine were obtained. The 39 target genes were uploaded to the string database for retrieval, and the species was defined as “Homo sapiens”, and the protein–protein interaction (PPI) network map was obtained. The node1, node2 and combined score information from the file exported from string database were imported into the software of Cytoscape 3.8.033,34 for visual analysis, and the network analysis results were obtained. Further, set the node size and color reflection (degree) value, the thickness of the edge reflects the combined score size, and finally get PPI network.
Identification of CI-Related Genes
GeneCards and DisGenNET databases31,32 were used to identify CI-related genes, and a total of 2929 results were identified in GeneCards database and 358 results were identified in DisGeNET database. After screening and deleting repetitive target genes, 2930 CI-related genes were obtained.
Identification of Related Targets of Ketamine and CI
CI-related genes (2930) and Ketamine targets (39) were uploaded to Venny 2.1.0 software to obtain the intersection map of Ketamine target and CI target, so as to obtain the potential target of Ketamine for CI treatment.
Screening of Key Target Proteins and GO and KEGG Analysis
The BisoGenet application in Cytoscape was used to build a PPI network of mutual targets between CI and ketamine. The key targets were screened by the scores of “degree”, “intermediate” and “intimacy” calculated by double genome. GO and KEGG enrichment analysis was performed in the Metascape database to identify the molecular function and systematic involvement of the target gene. The common targets of ketamine and CI were imported into Metascape database. Biological function (BP), molecular function (MF), cell composition (CC) and KEGG were checked respectively. Data with P < 0.05 were selected from BP, CC, MF and KEGG values, and then input into R software to draw GO analysis diagram and KEGG pathway enrichment analysis bubble diagram, so as to further explore the potential mechanism of ketamine in CI treatment.35,36
Molecular Docking
Three-dimensional (3D) structure of α-synuclein (SNCA; PDB ID: 3Q29), nitric oxide synthase 1 (NOS1; PDB ID: 6PNA) and muscarinic acetylcholine receptor M1 (CHRM1; PDB ID: 6WJC) was retrieved from RCSB PDB database.29 Using Autodock Vina software,37 water molecules were removed, proteins were separated, non-polar hydrogen was added, the Gasteiger charge of the structure was calculated, and saved as a PDBQT file. The two-dimensional (2D) and 3D structures of Ketamine (PubChem CID: 3821) were downloaded using the PubChem database.28 The 2D structures were converted to PDB format by Chem3D processing and saved as docking ligands in Autodock Vina software in PDBQT format. According to visually inspected and docking score, SNCA, NOS1 and CHRM1 were used as receptors, and ketamine was used as ligand. The conformation with the best affinity was selected as the final docking conformation, and PyMOL was used to analyze the docking results. The higher the absolute value of the binding free energy, the greater the stability of a protein–ligand complex.
Results
The Expected Target of Ketamine and Its Network
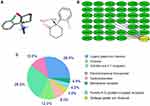
The 2D and 3D structures of Ketamine (PubChem CID: 3821) are shown in Figure 1A. For the purpose of investigating the target proteins of ketamine, we used SwissTargetPrediction, DrugBank and PubChem databases for prediction. Then, 39 target proteins were accessed by taking the intersection of the targets from these databases (Figure 1B). The three predominant types of these targets are ligand gated ion channels (28%), G-group protein coupled receptors (28%), and electrochemical transporters (12%) (Figure 1C). According to PPI network (Figure 2A), the critical potential target proteins with more interaction (binding fraction) with other targets were screened by using Cytoscape software. The target proteins located in the center of PPI network are 5-hydroxytryptamine receptor 1A (HTR1A), dopamine D2 receptor (DRD2) and 5-hydroxytryptamine receptor 3A (HTR3A) (Figure 2B). The top 15 target genes were mapped in R software according to the interactive data of PPI (Figure 2C).
Analysis of Gene Topological Networks Related to CI and Ketamine
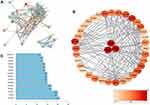
To obtain the critical genes in CI, we performed prediction using GeneCards and DisGeNET databases. The result indicated that a total of 2930 CI-related human genes were identified from the database. Using the Wayne map, there are 30 intersecting genes between ketamine targets and CI-related genes (Figure 3A). The information of 30 intersecting genes was shown in Figure 3B. In a PPI network, the degree center (DC) of a node is simply the number of edges it has. Betweenness centrality (BC) captures the position of a given node in relation to each other. The combination of DC and BC has been shown to be effective for screening reliable and important proteins.38 Subsequently, the PPI network of 30 target proteins was carried out by Cytoscape software and its BisoGenet application program. An extended network of 296 proteins associated with 30 target proteins was obtained. As shown in Figure 4A, the extended network has a total of 296 nodes (targets) and 1871 edges (interactions). In order to identify the most important nodes in these 296 proteins, the DC > 18 (twofold median) was used as the primary screening criteria, and 64 nodes and 520 edges were obtained (Figure 4B). Then, BC range of 0.548–38.573 was further screened. Finally, 24 related proteins and 149 correlations were obtained (Figure 4C), which may play an important role in the treatment of CI by ketamine. In addition, the first 20 of them were screened (Table 1).
 |
Table 1 Top 20 Potential Targets Associated with CI in Ketamine Treatment |
GO and KEGG Enrichment Analysis of 298 Related Genes
To explore the functions and related pathways of 298 target genes, we used GO and KEGG for macroscopic evaluation. According to CC analysis of GO, these proteins are mainly located in glutamatergic synapses, postsynaptic density, asymmetric synapses, synaptic membranes, and postsynaptic specialization (Figure 5A); in terms of MF, they mainly focus on ion channel binding, protein C-terminal binding, glutamate receptor binding, neurotransmitter receptor activity, and G protein coupled receptor binding (Figure 5B) Their BP mainly focus on divalent inorganic cation transport, divalent metal ion transport, calcium ion transport, regulation of trans-synaptic signal and regulation of chemical synaptic transmission (Figure 5C). KEGG pathway analysis further displayed that these proteins were mainly involved in day–night transmission, calcium signaling pathway, dopaminergic synapses, cholinergic synapses and glutamatergic synapses (Figure 5D and Table 2).
 |
Table 2 P Values of 10 Pathways and Pathways Related to Ketamine Involvement in Treatment of CI |
Molecular Docking Analysis of Ketamine and Its Potential Protein Targets
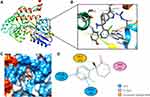
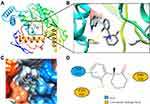
Molecular docking is a technique that simulates the interaction between small ligand molecules and receptor protein macromolecules, allowing the binding energy between the two counterparts to be calculated to predict their affinity. The analysis of the top 20 ketamine’s potential targets in Table 1 and the above key target proteins using molecular docking provides an intuitive explanation for the interaction between Ketamine and its CI-related potential target proteins. The top 10 targets for ketamine based on docking scores are shown in Table 3. Docking scores below 0 indicate spontaneous binding of two molecules, with smaller docking score leading to more stable conformations. According to visually inspected and docking score, we selected SNCA (PDB ID: 3Q29), NOS1 (PDB ID: 6PNA) and CHRM1 (PDB ID: 6WJC) with docking score of −9.9, −8.9 and −7.9 kcal/mol, respectively, for molecular docking with ketamine. The result confirmed that SNCA, NOS1 and CHRM1 are the most likely targets of ketamine, which have spatial adaptability, conventional hydrogen bound, Pi-alkyl interaction and Pi-Pi interaction are the main forms of interaction. As shown in Figure 6, ketamine interacted with amino acid residues TRP63, TRP341, TYR156 and GLU154 of SNCA, and formed hydrogen bonds with GLU154, Pi-alkyl interaction with TRP341, and Pi-Pi interaction with TRP63 and TYR156. As seen in Figure 7, ketamine formed hydrogen bonds with amino acid residues CYS420 and PRO570 of NOS1, and formed a Pi-Pi interaction with TRP414. In addition, amino acid residues TYR404 and TYR408, TYR381 and ASP105 of CHRM1 formed hydrogen bonds and Pi-Pi interaction with ketamine, respectively (Figure 8). The above results indicated that ketamine has good binding activity with SNCA, NOS1 and CHRM1.
 |
Table 3 Ketamine’s Top 10 Targets Based on Docking Scores |
Discussion
Ketamine has been used in medicine for more than 50 years, but its mechanism is still not well understood. It has a wide range of functions and complex pharmacological effects. Clinically, ketamine is applied as an analgesic, antidepressant, anti-inflammatory drug, dissociative anesthetic, local anesthetic, neuroprotective agent, psychotic, and even neurotoxins.10 In the past, it was mainly used in anesthesia, analgesia and antidepressant therapy. Now, ketamine has been shown to have improved behavioral and neuroprotective effects in animal models of CI.39 According to our network pharmacology results, SNCA, NOS1 and CHRM1 may play a key role in the treatment of Ci by ketamine.
Target SNCA
SNCA is a neuronal protein that plays multiple roles in synaptic activity, such as regulating synaptic vesicle transport and subsequent neurotransmitter release.40 Mechanistically, SNCA regulates dopamine transmission by increasing the release of Ca2+ in the middle of the microregion, which is also a major component of the Louis body. The aggregation of the protein in the cell represents a histological marker of Parkinson’s disease.41 In rodents suffering from focal ischemia, SNCA gene knockdown significantly diminished the induction of Drp1, 3-nitrotyrosine phosphate, caspase-3 and LC-3II/I after ischemia. These results indicated that it can induce mitochondrial fragmentation, oxidative stress, and autophagy, thus promoting neuronal cell death. Therefore, it plays a key role in ischemic brain injury and is a potential therapeutic target for reducing brain injury after stroke.42 If the expression of SNCA protein is stopped in the early stage after CI, brain injury can be minimized and a better functional outcome can be guided.43 Inhibition of SNCA gene expression in the animal model of CI can significantly decrease lesion volume, enhance exercise and facilitate cognitive function recovery.44 During the whole CI, the ablation of SNCA gene can increase the formation of prostaglandins in the brain, which have a protective effect against CI. Hence, reducing the level of SNCA can effectively reduce the post-ischemic brain injury.45 Notably, no studies have reported the progression of ketamine involvement in CI through regulation of SNCA. In this study, the network pharmacology studies we have done suggests that ketamine can bind tightly to SNCA firmly, inhibit protein function, reduce neuronal apoptosis, protect post-ischemic brain cells, and facilitate recovery of brain function.
Target NOS1
It has been demonstrated that activation of c-Jun N-terminal kinase 1/2 (JNK1/2) is a signaling pathway for early neuronal death in CI. This is because the expression of neuronal nitric oxide synthase (nNOS) is increased and more NO is produced.46 NO, as a free radical, can directly destroy protein structure, disrupt mitochondrial function, and inspire apoptosis.47 Previous studies disclosed that transient focal ischemia increases endothelial-type NOS1 in the cerebral vasculature.48 Therefore, inhibition of NOS1 (neuronal NOS) activity can alleviate cell death in CI.49 According to previous studies, intrathecal ketamine can significantly suppress the expression of NOS1 in the dorsal horn of the rat's spinal cord.50 Ketamine has been shown to be involved in the development of disorders such as schizophrenia and depression by regulating the expression of NOS1.51–53 Our molecular docking simulation results indicated that ketamine has strong docking ability with NOS1. It can effectively inhibit the activity of NOS1, which in turn reduces the production of NO, thus avoiding the damage to neurons by oxidative stress. Moreover, CI-reperfusion injury is associated to the calcium influx mediated by NMDA receptor, which activates nNOS and thus induces NO production.54 Ketamine can simultaneously inactivate NMDA receptor and NOS1, which should be one of the mechanisms by which ketamine can prevent CI injury.55
Target CHRM1
Ketamine can significantly inhibit CHRM1 and muscarinic signal. This effect may explain some anticholinergic effects of ketamine, including central (effect on memory and consciousness) and peripheral (significant enhancement of sympathetic excitability, bronchiectasis, mydriasis, etc).56 Previous studies have demonstrated that the absence of CHRM in microglia affects CI injury.57 T-type calcium channel enhancer SAK3 inhibits neuronal death after transient CI via CHRM1 stimulation.58 Previous studies have revealed that ketamine can reduce the need for variable force drug support in patients with septic shock, and its effect may be related to the inhibition of CHRM1.59 Ketamine is the only intravenous anesthetic that can stimulate brain function and leads to a 50% increase in cerebral blood flow.60 At the same time, it can reduce catecholamine reuptake, increase mean arterial pressure, and make hemodynamics more stable.61 It can also increase heart rate and myocardial contractility by stimulating sympathetic nerve, and its cardiovascular excitation response can improve cerebral perfusion and has neuroprotective properties.62 Based on this, ketamine is helpful for the recovery of blood perfusion of tissue cells after CI by regulated CHRM1.
More Relevant Mechanisms
We conclude through the network pharmacology that SNCA, NOS1, and CHRM1 are the key targets of ketamine treatment CI. Molecular docking simulation has the best and most stable binding force, which is consistent with the previous research results. Ketamine even has anti-inflammatory properties, which can skew macrophages to an M2-like phenotype.63 Furthermore, ketamine can inhibit the activation of leukocytes in inflammatory response and reduce the production of inflammatory cytokines TNF-α and interleukin-6, thereby reducing the neurological damage caused by the inflammatory response.64 Previous studies have confirmed that subanesthetic doses of ketamine can activate autophagy activity and protect neurons in a mouse model of Parkinson’s disease.65 The expression of c-Jun protein in the hippocampus of mice can be regulated to protect the mice from CI and reperfusion injury.66 This is in line with the results we have analyzed from KEGG and GO. According to CC analysis of GO, 296 related target genes involved in ketamine treatment CI were mainly located in glutamate synapses, postsynaptic density, asymmetric synapses, synaptic membrane and postsynaptic specialization. The analysis of KEGG pathway exhibited that these proteins mainly involved in the day and night transmission, calcium signaling pathway, dopamine synapse, cholinergic synapse and glutamate synapse, and almost all involved in the information transmission of nervous system.
Limitations and Future Directions
Ketamine has unique pharmacological advantages and multi-target characteristics that distinguish it from other traditional anesthetics. It has a wide range of clinical prospects and has already exceeded the scope of general anesthetics. Sub-anaesthetic dose of ketamine is now more widely used in the treatment of chronic pain, Alzheimer’s disease, depression and CI. Nevertheless, it is not limited to this. In the future, there are more application directions to be explored. However, there is still lack of large sample, long-term and repeated clinical evidence of ketamine in the prevention of nerve injury after CI. The neurotoxic effect of ketamine is also a major obstacle limiting its clinical application, and the problem of addiction must also be taken seriously. Further research is needed to determine the selection criteria, optimal scheme and dose selection, and minimize adverse reactions to improve its clinical efficacy and safety.
Conclusion
In summary, we explored the multi-target mechanism of ketamine to treat CI through network pharmacology. The findings indicated SNCA, NOS1 and CHRM1 are the most likely targets for ketamine on treating CI. This article uses network pharmacology and molecular docking simulation methods to explore the possible mechanism of ketamine for CI treatment, which will help facilitate its clinical application in more directions.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This research was supported by a grant from The National Natural Science Foundation of China (grant No.81760248, 81960250). The authors also would like to thank the Medical University of Kunming (Kunming, China) for its technical support.
Funding
This work was supported by the National Natural Science Foundation of China (grant No.81760248, 81960250).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kim J, Thayabaranathan T, Donnan GA, et al. Global stroke statistics 2019. Int J Stroke. 2020;15(8):819–838. doi:10.1177/1747493020909545
2. Wang W, Jiang B, Sun H, et al. Prevalence, incidence, and mortality of stroke in china: results from a nationwide population-based survey of 4,80,687 adults. Circulation. 2017;135(8):759–771. doi:10.1161/CIRCULATIONAHA.116.025250
3. Ekker MS, Verhoeven JI, Vaartjes I, et al. Association of stroke among adults aged 18 to 49 years with long-term mortality. JAMA. 2019;321(21):2113–2123. doi:10.1001/jama.2019.6560
4. Thrift AG, Thayabaranathan T, Howard G, et al. Global stroke statistics. Int J Stroke. 2017;12(1):13–32. doi:10.1177/1747493016676285
5. Yenari MA, Han HS. Neuroprotective mechanisms of hypothermia in brain ischaemia. Nat Rev Neurosci. 2012;13(4):267–278. doi:10.1038/nrn3174
6. Li W, Ye A, Ao L, et al. Protective mechanism and treatment of neurogenesis in cerebral ischemia. Neurochem Res. 2020;45(10):2258–2277. doi:10.1007/s11064-020-03092-1
7. Chamorro A, Dirnagl U, Urra X, et al. Neuroprotection in acute stroke: targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016;15(8):869–881. doi:10.1016/S1474-4422(16)00114-9
8. Mallet RT, Ryou MG. Erythropoietin: endogenous protection of ischemic brain. Vitam Horm. 2017;105:197–232.
9. Suzuki K, Nosyreva E, Hunt KW, et al. Effects of a ketamine metabolite on synaptic NMDAR function. Nature. 2017;546(7659):E1–E3. doi:10.1038/nature22084
10. Zanos P, Moaddel R, Morris PJ, et al. Ketamine and ketamine metabolite pharmacology: insights into therapeutic mechanisms. Pharmacol Rev. 2018;70(3):621–660. doi:10.1124/pr.117.015198
11. Mion G, Villevieille T. Ketamine pharmacology: an update (pharmacodynamics and molecular aspects, recent findings). CNS Neurosci Ther. 2013;19(6):370–380. doi:10.1111/cns.12099
12. Swainson J, Thomas RK, Archer S, et al. Esketamine for treatment resistant depression. Expert Rev Neurother. 2019;19(10):899–911. doi:10.1080/14737175.2019.1640604
13. Malinow R. Depression: ketamine steps out of the darkness. Nature. 2016;533(7604):477–478. doi:10.1038/nature17897
14. Tang SH, Yu JG, Li JJ, et al. Neuroprotective effect of ketamine on acute spinal cord injury in rats. Genet Mol Res. 2015;14(2):3551–3556. doi:10.4238/2015.April.17.4
15. De Kock M, Loix S, Lavand’homme P. Ketamine and peripheral inflammation. CNS Neurosci Ther. 2013;19(6):403–410. doi:10.1111/cns.12104
16. Wang CQ, Ye Y, Chen F, et al. Posttraumatic administration of a sub-anesthetic dose of ketamine exerts neuroprotection via attenuating inflammation and autophagy. Neuroscience. 2017;343:30–38. doi:10.1016/j.neuroscience.2016.11.029
17. Luo TT, Lu Y, Yan SK, et al. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med. 2020;26(1):72–80. doi:10.1007/s11655-019-3064-0
18. Hsin KY, Matsuoka Y, Asai Y, et al. systemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Res. 2016;44(W1):W507–W513. doi:10.1093/nar/gkw335
19. Ding M, Ma W, Wang X, et al. A network pharmacology integrated pharmacokinetics strategy for uncovering pharmacological mechanism of compounds absorbed into the blood of Dan-Lou tablet on coronary heart disease. J Ethnopharmacol. 2019;242:112055. doi:10.1016/j.jep.2019.112055
20. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi:10.1038/nchembio.118
21. Jacunski A, Tatonetti NP. Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Ther. 2013;94(6):659–669. doi:10.1038/clpt.2013.168
22. Kibble M, Saarinen N, Tang J, et al. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32(8):1249–1266. doi:10.1039/C5NP00005J
23. Zhao S, Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu Rev Pharmacol Toxicol. 2012;52:505–521. doi:10.1146/annurev-pharmtox-010611-134520
24. Casas AI, Hassan AA, Larsen SJ, et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc Natl Acad Sci U S A. 2019;116(14):7129–7136. doi:10.1073/pnas.1820799116
25. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
26. Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. doi:10.1093/nar/gku293
27. Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi:10.1093/nar/gkj067
28. Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–D1395. doi:10.1093/nar/gkaa971
29. Karuppasamy MP, Venkateswaran S, Subbiah P. PDB-2-PBv3.0: an updated protein block database. J Bioinform Comput Biol. 2020;18(2):2050009. doi:10.1142/S0219720020500092
30. Sterling T, Irwin JJ. ZINC 15–ligand discovery for everyone. J Chem Inf Model. 2015;55(11):2324–2337. doi:10.1021/acs.jcim.5b00559
31. Fishilevich S, Nudel R, Rappaport N, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017;2017. doi:10.1093/database/bax028
32. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi:10.1093/nar/gkw943
33. Otasek D, Morris JH, Bouças J, et al. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi:10.1186/s13059-019-1758-4
34. Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi:10.1038/nmeth.2212
35. Martin A, Ochagavia ME, Rabasa LC, et al. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinform. 2010;11:91. doi:10.1186/1471-2105-11-91
36. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
37. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
38. Wang J, Peng W, Wu FX. Computational approaches to predicting essential proteins: a survey. Proteom Clin Appl. 2013;7(1–2):181–192. doi:10.1002/prca.201200068
39. Wang R, Zhang Z, Kumar M, et al. Neuroprotective potential of ketamine prevents developing brain structure impairment and alteration of neurocognitive function induced via isoflurane through the PI3K/AKT/GSK-3β pathway. Drug Des Devel Ther. 2019;13:501–512. doi:10.2147/DDDT.S188636
40. Ulusoy A, Di Monte DA. α-Synuclein elevation in human neurodegenerative diseases: experimental, pathogenetic, and therapeutic implications. Mol Neurobiol. 2013;47(2):484–494. doi:10.1007/s12035-012-8329-y
41. Henderson MX, Trojanowski JQ, Lee VM. α-Synuclein pathology in Parkinson’s disease and related α-synucleinopathies. Neurosci Lett. 2019;709:134316. doi:10.1016/j.neulet.2019.134316
42. Kim T, Mehta SL, Kaimal B, et al. Poststroke induction of α-synuclein mediates ischemic brain damage. J Neurosci. 2016;36(26):7055–7065. doi:10.1523/JNEUROSCI.1241-16.2016
43. Chelluboina B, Kim T, Mehta SL, et al. Impact of age and sex on α-Syn (α-Synuclein) knockdown-mediated poststroke recovery. Stroke. 2020;51(10):3138–3141. doi:10.1161/STROKEAHA.120.028978
44. Kim T, Mehta SL, Morris-Blanco KC, et al. The microRNA miR-7a-5p ameliorates ischemic brain damage by repressing α-synuclein. Sci Signal. 2018;11:560. doi:10.1126/scisignal.aat4285
45. Kim T, Vemuganti R. Mechanisms of Parkinson’s disease-related proteins in mediating secondary brain damage after cerebral ischemia. J Cereb Blood Flow Metab. 2017;37(6):1910–1926. doi:10.1177/0271678X17694186
46. Yu HM, Xu J, Li C, et al. Coupling between neuronal nitric oxide synthase and glutamate receptor 6-mediated c-Jun N-terminal kinase signaling pathway via S-nitrosylation contributes to ischemia neuronal death. Neuroscience. 2008;155(4):1120–1132. doi:10.1016/j.neuroscience.2008.03.061
47. Zhou L, Li F, Xu HB, et al. Treatment of cerebral ischemia by disrupting ischemia-induced interaction of nNOS with PSD-95. Nat Med. 2010;16(12):1439–1443. doi:10.1038/nm.2245
48. Veltkamp R, Rajapakse N, Robins G, et al. Transient focal ischemia increases endothelial nitric oxide synthase in cerebral blood vessels. Stroke. 2002;33(11):2704–2710. doi:10.1161/01.STR.0000033132.85123.6A
49. Zhang CZ, Dong L, Mu FH, et al. [Progress in the studies on neuronal nitric oxide synthase inhibitors]. Yao Xue Xue Bao. 2014;49(6):781–788. Chinese.
50. Yang Y, Guo QL, Zou WY, et al. [Effect of intrathecal ketamine on the expression of nNOS in spinal dorsal horn in rats with formalin pain]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2006;31(5):747–751. Chinese.
51. Pereira VS, Suavinha A, Wegener G, et al. Prelimbic neuronal nitric oxide synthase inhibition exerts antidepressant-like effects independently of BDNF signalling cascades. Acta Neuropsychiatr. 2019;31(3):143–150. doi:10.1017/neu.2018.39
52. Keilhoff G, Becker A, Grecksch G, et al. Repeated application of ketamine to rats induces changes in the hippocampal expression of parvalbumin, neuronal nitric oxide synthase and cFOS similar to those found in human schizophrenia. Neuroscience. 2004;126(3):591–598. doi:10.1016/j.neuroscience.2004.03.039
53. Chen J, Zhang M, Zhou C, et al. Association analysis of neuronal nitric oxide synthase 1 gene polymorphism with psychopathological symptoms in chronic ketamine users. Front Psychiatry. 2020;11:580771. doi:10.3389/fpsyt.2020.580771
54. Hsu YC, Chang YC, Lin YC, et al. Cerebral microvascular damage occurs early after hypoxia-ischemia via nNOS activation in the neonatal brain. J Cereb Blood Flow Metab. 2014;34(4):668–676. doi:10.1038/jcbfm.2013.244
55. Kokkinou M, Ashok AH, Howes OD. The effects of ketamine on dopaminergic function: meta-analysis and review of the implications for neuropsychiatric disorders. Mol Psychiatry. 2018;23(1):59–69. doi:10.1038/mp.2017.190
56. Durieux ME. Inhibition by ketamine of muscarinic acetylcholine receptor function. Anesth Analg. 1995;81(1):57–62. doi:10.1097/00000539-199507000-00012
57. Costa A, Haage V, Yang S, et al. Deletion of muscarinic acetylcholine receptor 3 in microglia impacts brain ischemic injury. Brain Behav Immun. 2021;91:89–104. doi:10.1016/j.bbi.2020.09.008
58. Yabuki Y, Jing X, The FK. T-type calcium channel enhancer SAK3 inhibits neuronal death following transient brain ischemia via nicotinic acetylcholine receptor stimulation. Neurochem Int. 2017;108:272–281. doi:10.1016/j.neuint.2017.04.015
59. Durieux ME, Nietgen GW. Synergistic inhibition of muscarinic signaling by ketamine stereoisomers and the preservative benzethonium chloride. Anesthesiology. 1997;86(6):1326–1333. doi:10.1097/00000542-199706000-00014
60. Bowles ED, Gold ME. Rethinking the paradigm: evaluation of ketamine as a neurosurgical anesthetic. Aana j. 2012;80(6):445–452.
61. Hoffman WE, Pelligrino D, Werner C, et al. Ketamine decreases plasma catecholamines and improves outcome from incomplete cerebral ischemia in rats. Anesthesiology. 1992;76(5):755–762. doi:10.1097/00000542-199205000-00014
62. Huang M, Watso JC, Moralez G, et al. Low-dose ketamine affects blood pressure, but not muscle sympathetic nerve activity, during progressive central hypovolemia without altering tolerance. J Physiol. 2020;598(24):5661–5672. doi:10.1113/JP280491
63. Nowak W, Grendas LN, Sanmarco LM, et al. Pro-inflammatory monocyte profile in patients with major depressive disorder and suicide behaviour and how ketamine induces anti-inflammatory M2 macrophages by NMDAR and mTOR. EBioMed. 2019;50:290–305. doi:10.1016/j.ebiom.2019.10.063
64. Bartoc C, Frumento RJ, Jalbout M, et al. A randomized, double-blind, placebo-controlled study assessing the anti-inflammatory effects of ketamine in cardiac surgical patients. J Cardiothorac Vasc Anesth. 2006;20(2):217–222. doi:10.1053/j.jvca.2005.12.005
65. Fan JC, Song JJ, Wang Y, et al. Neuron-protective effect of subanesthestic-dosage ketamine on mice of Parkinson’s disease. Asian Pac J Trop Med. 2017;10(10):1007–1010. doi:10.1016/j.apjtm.2017.09.014
66. Xiao F, Xiong L, Wang Q, et al. Ketamine inhibits c-Jun protein expression in mouse hippocampus following cerebral ischemia/reperfusion injury. Neural Regen Res. 2012;7(11):833–836. doi:10.3969/j.issn.1673-5374.2012.11.006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.