Back to Journals » International Journal of Nanomedicine » Volume 16
Exosomes Promote the Transition of Androgen-Dependent Prostate Cancer Cells into Androgen-Independent Manner Through Up-Regulating the Heme Oxygenase-1
Authors Zhang Y , Chen B, Xu N, Xu P, Lin W, Liu C, Huang P
Received 13 September 2020
Accepted for publication 28 December 2020
Published 12 January 2021 Volume 2021:16 Pages 315—327
DOI https://doi.org/10.2147/IJN.S281710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Yiming Zhang,1,2,* Binshen Chen,1,2,* Naijin Xu,3 Peng Xu,1,2 Wenfeng Lin,1– 3 Chunxiao Liu,1,2 Peng Huang1– 3
1Department of Urology, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 2Guangzhou Key Laboratory of Inflammatory and Immune Diseases, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 3Department of Urology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan
*These authors contributed equally to this work
Correspondence: Peng Huang; Chunxiao Liu
Department of Urology, Zhujiang Hospital, Southern Medical University, Guangzhou 510282, People’s Republic of China
Tel +86-20-62782726
Fax +86-20-62782725
Email [email protected]; [email protected]
Background: Castration-resistant prostate cancer (CRPC) is still considered incurable, even though the mechanisms of CRPC had been extensively researched. Studies have demonstrated that exosomes in the tumor microenvironment contribute to prostate cancer development and progression. However, the role of exosomes in the process of CRPC progression has not yet been determined.
Methods: Co-culturing and exosome treatment assays combined with in vitro and in vivo assays were performed to determine the function of exosomes in the transformation of androgen-dependent prostate cancer (ADPC) cells into androgen-independent cells. Then, the mRNA expression profiles of ADPC cells and ADPC cells co-cultured with androgen-independent prostate cancer (AIPC) cell-derived exosomes were studied using microarrays. After silencing the expression of heme oxygenase-1 (HMOX1), Western blotting, quantitative real-time PCR, immunohistochemistry (IHC) studies, and MTS assay were used to confirm the mechanisms of exosome participation in CRPC progression.
Results: The results showed that ADPC cells acquired tolerance for androgen deprivation due to the exosome-mediated communication between cells. AIPC cell-derived exosomes promoted the transformation of ADPC cells into androgen-independent cells in vivo and in vitro. Microarray analysis revealed that HMOX1 in ADPC cells was up-regulated after treatment with AIPC cell-derived exosomes. Further results showed that HMOX1 is overexpressed in human AIPC specimens and protects ADPC cells from androgen deprivation.
Conclusions: Our findings revealed that exosomes contribute to CRPC progression via promoting the transition of prostate cancer cells into an androgen-independent growth stage by activating HMOX1.
Keywords: prostate cancer, castration resistance, exosomes, heme oxygenase-1
Introduction
Prostate cancer is the second leading cause of cancer-related death in men.1 The treatment of prostate cancer includes radical resection, radiotherapy, local treatment (cryotherapy, radiofrequency ablation and high-energy focused ultrasound), endocrine therapy and chemotherapy.2 While endocrine therapy is effective in the early stage of metastasis, significantly inhibits the progression of metastatic prostate cancer and greatly improves the 5-year survival rate and quality of life of patients, most patients eventually develop castration-resistant prostate cancer (CRPC).2 Therefore, it is necessary to explore the mechanism and process of the development of hormone-independent prostate cancer.
Androgen resistance may be due to apoptosis of androgen-dependent prostate cancer (ADPC) cells during endocrine therapy. Prostate cancer stem cells are hormone independent, have a selective growth advantage, gradually develop from a small population to large population, and the last surviving cells are not androgen-dependent.3 A study showed that Bcl-2 could not be detected in 13 of 19 androgen-dependent prostate cancer samples, but Bcl-2 was strongly positive in androgen-independent prostate cancer samples. This finding suggested that the abnormal expression of Bcl-2 may be related to androgen resistance.4 Studies have also shown that androgen receptor (AR) sensitivity is significantly increased in more than 30% of androgen-independent prostate cancer tissues, and thus low levels of androgen can activate AR and promote the proliferation of prostate cancer cells.5–7AR is located on the X chromosome, and a study showed that a point mutation in the 877 amino acid of an AR transformed the inhibitory effect of flutamide on LNCAP prostate cancer cells into a growth-promoting effect.8 Although current studies have shown that the transition from androgen-dependent to androgen-independent prostate cancer may be closely related to these mechanisms, the process of this transition cannot be fully explained by the mechanisms mentioned above. Therefore, we speculate that there may be other more complex mechanisms involved in the transition from androgen-dependent to androgen-independent prostate cancer.
Exosomes are small membrane vesicles of endocytic origin, which are rich in cytoplasmic proteins, lipids, and nucleic acids. They play an important role in intercellular communications, especially in the tumor microenvironment.9 Many studies have shown that exosomes are involved in the development of prostate cancer. Corcoran et al found that docetaxel-resistant prostate cancer cells (22RV1RD and DU145RC) confer drug-resistance to 22RV1, DU145, and LNCAP exosomal MDR-1/P-gp transfer.10 Exosomes secreted by prostate cancer cells can down-regulate the expression of NKG2D on the surface of natural killer cells and CD8+ T cells, and NKG2D is the receptor that activates cytotoxicity. The inactivation of NKG2D leads to immunosuppression and immune escape of tumor cells. The expression of NKG2D on the surface of natural killer cells and CD8+ T cells is significantly decreased in CRPC patients.11 Exosomes secreted by prostate cancer cells can promote the differentiation of bone marrow mesenchymal stem cells into myofibroblasts, which are capable of angiogenesis and promote tumor proliferation and invasion.12
Many studies have shown that the tumor microenvironment plays an important role in the malignant transformation of cancers, including prostate cancer (proliferation, invasion, migration, immunosuppression and chemotherapy resistance).9 Therefore, we also speculate that the tumor microenvironment may play a sustained and important role in the whole process of androgen resistance. Exosomes play an important role in cell–cell interactions in the tumor microenvironment, and recent studies have found that exosomes rich in mir-1290 and mir-375 are closely related to the survival and prognosis of hormone-resistant prostate cancer patients.13 Therefore, we speculate that exosomes may also play a role in the emergence of hormone resistance, ie, cell populations that have become androgen-resistant can accelerate the whole process of androgen resistance by producing exosomes that act on androgen-dependent cell populations causing them to become androgen-independent prostate cancer cells.
The present study aimed to ascertain whether exosomes are involved in the transition of human androgen‑dependent prostate cancer cells into androgen-independent cells. Our data revealed that androgen-independent prostate cancer cells transform ADPC cells into androgen-independent cells via delivery of exosomes and overexpression of HMOX1.
Materials and Methods
Cell Lines and Culture
The prostate cancer cell lines LNCAP and PC-3 were supplied by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and had been authenticated by Short Tandem Repeat (STR) profiling. These cells were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and penicillin (100 units/mL) and streptomycin (100 mg/mL) (Life Technologies) at 37°C with 5% CO2. Hemin (HMOX1 agonist) was purchased from Sigma-Aldrich (USA), and was diluted to a final concentration 50 μM.
For androgen ablation experiments, and prior to treatment with dihydrotestosterone (DHT), LNCAP cells were cultured in RPMI-1640 (Gibco, USA) supplemented with 10% charcoal/dextran-treated FBS (HyClone, USA). For exosomal studies, exosome-depleted FBS was purchased from System Biosciences, USA (catalog: EXO-FBS-250A-1).
Isolation of Exosomes
This study used a modified and efficient method for the extraction of exosomes that was described by Zhu.14 Briefly, the prostate cancer cells were plated in 20 mL of RPMI-1640 containing 10% FBS, 100 units/mL penicillin, and 100 mg/mL streptomycin on 15cm dishes. When the cells reached approximately 80% confluence, they were washed 3 times with phosphate-buffered saline (1×PBS) and were then cultured in exosome-depleted RPMI-1640 medium. After 48h, the culture supernatant was collected and centrifuged at 3000×g for 15 min at 4°C. Then, an Ultracel centrifugal filter device with a 3 kD molecular weight cutoff (Millipore, USA) was used to concentrate the supernatant via centrifugation in a swing-out rotor and 4000×g at 4°C. The concentrated supernatant was further treated with ExoQuick-TC (System Biosciences, USA) for final exosome isolation according to the manufacturer’s protocol. The exosome pellet was dissolved in 500 μL of 1×PBS.
Transmission Electron Microscopy (TEM)
The morphology of purified exosomes was examined by TEM. In brief, exosomes were loaded onto 200 mesh copper grids coated with Formvar-carbon and incubated for 2 min at room temperature. Grids were stained with 20μL 2.0% phosphotungstic acid and allowed to dry. A transmission electron microscope (HITACHI S-900, Japan) operated at 80 kV was used to view samples.
cDNA Microarray Analysis
Gene expression profiling was performed using an Affymetrix Gene Chip® PrimeView™ Human Gene Expression Array, which contains 36,000 probe sets representing 20,000 well-characterized human genes (Affymetrix, Santa Clara, CA). Total RNA was isolated from duplicate cultures using the TRIzol method. After passing a quality control measurement, RNA was amplified and labeled using the Affymetrix 3ʹ IVT Express kit and hybridized to the array for 16 h. This was followed by washing and staining using the Affymetrix Fluidics Station 450/250. Arrays were scanned using an Affymetrix 3000 7G plus instrument. Affymetrix GeneChip Command Console (AGCC) software was used to generate.cel files. Data were imported into Agilent Gene Spring GX software for further analysis. Unsupervised hierarchical clustering analysis was performed using Cluster 3.0 software (Stanford University, USA). The row dendrogram was generated using Ward’s clustering method with a half square Euclidean distance measure. The column dendrogram was generated using the single linkage clustering method and a Euclidean distance measure. Gene ontology analysis was based on David 6.7 (http://david.abcc.ncifcrf.gov/home.jsp).
The transcriptome data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE145786.
Co-Culture of LNCAP Cells with PC-3 Cells
Using a Transwell co-culture system, LNCAP cells (1×105) were seeded on a 0.4-μm Transwell pore-sized insert (Corning) and were co-cultured with PC-3 cells at the density of 4×105 cells/well in 6-well plates for 1 week. If the PC-3 cells of lower chamber had reached 70% to 80% confluence, the upper insert was transferred to another well with 4×105 PC-3 cells for further co-culturing.
Cell Proliferation Assay
For cell proliferation assays, cells were seeded in 96-well plates at 2×103 cells/well at a final volume of 100 µL and incubated overnight. The viability of cells was determined with a CellTiter 96 non-radioactive cell proliferation assay (MTS) (Promega BioSciences, USA) following the manufacturer’s protocol.
Cell Cycle Analysis
For cell cycle analysis, cells were fixed with 70% ethanol in PBS at 4°C overnight and then treated with ribonuclease to digest RNA and stained with 50 µg/mL of propidium iodide (PI). Cell cycle analysis was then performed by FACSCaliber BD flow cytometry.
In vivo Tumorigenicity Assay
Male NOD/SCID mice 4 to 6 weeks of age were purchased from Beijing Vital River Laboratories (Beijing, China). All mouse experiments were approved by the local ethics committee of Zhujiang Hospital. Prior to the in vivo androgen ablation experiments, mice were surgically castrated. For tumor growth assays, LNCAP cells were first treated with PC-3 derived exosomes (100μg/mL) every 2 days for a total of 14 days, while the control group was treated with PBS. Cells were then subcutaneously injected into the dorsal flank of intact or castrated NOD/SCID mice. The mice were killed 45 days after the injection of cells, and tumors were harvested.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated with TRIzol reagent (Invitrogen), and cDNA was synthesized with a reverse-transcription kit (Takara, Japan). Quantitative analysis was performed using the LightCycler® 480 SYBR Green I Master (Roche, Switzerland) on an Eco Real-Time PCR System (Illumina, USA) according to the manufacturer’s instructions. The relative mRNA expression was calculated using the 2−ΔΔCt normalized to GAPDH. Primer sequences used are shown in Table 1.
 |
Table 1 List of the Primer Sequences for PCR Studies |
Western Blotting
Cells were lysed in RIPA buffer (Beyotime, China) containing protease inhibitors (Complete Mini Ethylene Diamine Tetraacetic Acid-Free Protease Inhibitor Cocktail, Roche). The protein lysates were centrifuged, and the supernatants were collected for protein quantification with a Bicinchoninic Acid assay kit (Pierce, USA). Protein lysates were resolved with 8% to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Beyotime, China), and were electrophoretically transferred to nitrocellulose filter membranes (Millipore). The membranes were then probed with antibodies against prostate-specific antigen (PSA) (1:1000; Cell Signaling Technology), AR (1:1000; Cell Signaling Technology), TSG101 (1:1000; System Biosciences), HSP70 (1:1500; System Biosciences), Alix (1:1000; System Biosciences) and HMOX1 (1:1000; Abcam). GAPDH (1:800; Cell Signaling Technology) and β-actin (1:1000; Cell Signaling Technology) were used as loading controls. After incubation with horseradish peroxidase (HRP)–conjugated goat anti-rabbit immunoglobulin G or HRP-conjugated goat anti-mouse immunoglobulin G secondary antibodies, the proteins were observed with the ChemiDoc XRS+ system (Bio-Rad, USA).
Immunohistochemistry
Immunohistochemistry (IHC) studies were performed on paraffin-embedded tissue sections from ADPC and androgen-independent prostate cancer (AIPC) tissues. Immunostaining was performed with an anti-HMOX1 antibody (Abcam) at 4°Cwith the EnVision HRP rabbit detection system (Dako, CA). Tissue sections were then treated with 3% hydrogen peroxide for 10 min. The primary antibody was diluted (1:300) with a background reducing diluent (Dako) and was incubated at 4°C overnight. Slides were incubated with the EnVision reagent for 30 min. The slides were then incubated with 3.30-diaminobenzidine for 3 min, counterstained with Meyer’s hematoxylin, and mounted. The samples were rinsed with a PBS buffer between each step. The percent of cells staining positive in each sample was determined in sections by counting the number of positive cells in 3 separate random sections containing 100 cells at a magnification of 200×.
Small Interfering RNA (siRNA) Transfection
Effective siRNAs targeting human HMOX1 were purchased from RiboBio (Guangzhou, China). siRNAs were transfected with RNAiMAX (Invitrogen). Subsequently, qRT-PCR and Western blotting analysis were performed to verify changes in expression.
Bioinformatics Analysis
To determine the expression pattern of HMOX1 in prostate cancer, the datasets in the Oncomine Cancer Microarray database (https://www.oncomine.org) were used.
Statistical Analysis
SPSS version 20.0 software (SPSS Inc. Chicago, USA) was used to perform all statistical analyses. Results were reported as the mean ± standard deviation of 3 independent experiments, and the significance of Differences among 3 groups were examined using one-way analysis of variance (ANOVA), while Student’s t-test was used to examine differences between 2 groups. A value of P < 0.05 was considered to be statistically significant.
Results
Induction of Resistance to Androgen Deprivation in LNCAP Cells Co-Cultured with PC-3 Cells
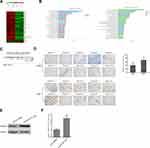
Due to the long process of ADPC becoming AIPC, there may be an “intermediate stage” (ADPC and AIPC cells coexist) before the final development of CRPC. Hence, our study intended to investigate the interaction between these 2 populations of cells using an ADPC cell model (LNCAP) and an AIPC cell model (PC-3). First, LNCAP cells were co-cultured with PC-3 cells in Transwell system for 1 week, and we observed that the morphology of LNCAP cells changed from epithelial-like to mesenchymal-like. qRT-PCR analysis showed that the marker of mesenchymal cells, N-cadherin, was up-regulated after LNCAP cells were co-cultured with PC-3 cells, while the epithelial marker E-cadherin was down-regulated (Supplementary Figure 1). The MTS assay results indicated that the proliferation of LNCAP cells in androgen-deprived medium was significantly increased after co-culturing (P<0.05), but there was no difference in when cells were cultured in general medium (Figure 1A). Flow cytometry was used to examine if the cell cycle of LNCAP cells was influenced by co-culturing with PC-3 cells, and the results showed that there was a higher rate of LNCAP cells in S-phase after co-culturing with PC-3 cells as compared with the control group under the castration condition (P<0.05), but no difference in normal condition (Figure 1B). Subsequent qRT-PCR analysis showed that AR mRNA expression and most AR-responsive genes (CDK1, CDK2 and GRBE1) in LNCAP cells were down-regulated after co-culturing (P<0.05) (Figure 1C), and the DHT effect on AR was attenuated (P<0.05) (Figure 1D). In addition, Western blotting confirmed that co-culturing LNCAP cells with PC-3 enhances AR-independent cell growth (Figure 1E).
Exosome Isolation and Identification
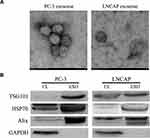
Cancer cells have been reported to secrete exosomes that function in an autocrine or paracrine manner to modulate tumor behavior.15 To determine whether PC-3 cells facilitate the emergence of castration resistance in LNCAP cells via secreted exosomes, we isolated exosomes from the culture supernatant of prostate cancer cells (LNCAP and PC-3). TEM showed membrane-limited particles that were homogeneous in appearance and ranged 50–100 nm in size (Figure 2A). Western blotting analysis showed the isolated particles expressed markers associated with exosomes (Alix, TSG101 and Hsp70) (Figure 2B). These results indicated that the particles had the characteristics of exosomes, and could be isolated in a consistent manner.
AIPC Cell-Derived Exosomes Promote Emergence of Castration Resistance in ADPC Cells
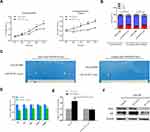
To test whether the AIPC cell-derived exosomes had the ability to confer castration resistance to ADPC cells, we incubated LNCAP cells with PC-3 derived exosomes (100 μg/mL) and conducted cell proliferation assays. As shown in Figure 3A, incubation of LNCAP cells with PC-3 exosomes caused a notable increase in cell growth in general or androgen-deprived medium (P<0.05). Flow cytometry showed that LNCAP cells exhibited a significantly higher rate of cells in S-phase after treatment with PC-3 exosomes compared with the control group, in both the normal condition and castration condition (Figure 3B; P<0.05).
Next, we confirmed the impact of PC-3 cell-derived exosomes on LNCAP tumor development in intact and castrated NOD/SCID mice. The results indicated that the tumorigenicity of LNCAP cells treated with PC-3 cell exosomes was significantly increased compared with normal LNCAP cells under the general condition (Figure 3C; P<0.05). Importantly, under the castration condition, normal LNCAP cells were unable to form tumors, whereas LNCAP cells treated with PC-3 exosomes were still able to develop tumors (Figure 3C). The tumor formation rates are summarized in Table 2.
 |
Table 2 Summary of Tumor Formation |
Western blotting and qRT-PCR were performed to examine AR signaling in LNCAP cells treated with PC-3 exosomes. AR mRNA expression of AR was significantly decreased in LNCAP cells treated with PC-3 cell exosomes as compared to untreated LNCAP cells (Figure 3D; P<0.05). In addition, the sensitivity of AR to androgen was reduced after treatment with PC-3 cell exosomes (Figure 3E; P<0.05). Western blotting showed that AR and PSA protein expression in LNCAP cells was significantly decreased after treatment with PC-3 cell exosomes (Figure 3F). These results indicate that AIPC cell population may cause the ADPC cell population to develop androgen resistance by the secretion of exosomes.
HMOX1 is a Possible Target of AIPC-Derived Exosomes in ADPC Cells
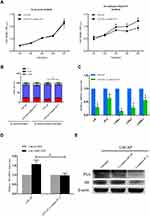
To explore the mechanisms of AIPC exosomes-induced castration resistance, we performed comprehensive gene expression profiling on LNCAP cells and LNCAP cells treated with PC-3 exosomes using Affymetrix microarray technology. An overview of the mRNA expression profiles is provided in Figure 4A. Based on the criteria of an adjusted value of P<0.05 and a minimum 1.5-fold change, 72 transcripts were found to be significantly affected, with 67 transcripts increased and 5 transcripts decreased. The 10 most affected genes are shown in Table 3. As shown in Figure 4B, the most significantly affected categories of genes were those involved in the inflammatory response, and the KEGG pathway analysis suggested most affected genes were involved in the activation of C3 and C5.
 |
Table 3 The 10 Most Affected Genes in PC-3 Exosomes-Treated LNCAP Cells |
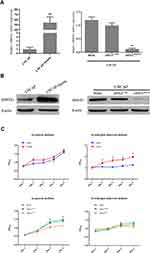
The microarray results suggested that PC-3 exosomes induced up-regulation of HMOX1, a member of the heat shock protein family, in LNCAP cells. HMOX1 modulates cell-adaptive responses to oxidative stress and has been previously implicated in the progression of hormone-refractory prostate cancer.16 To assess the role of HMOX1 in prostate cancer, we first analyzed 5 independent microarray datasets from the Oncomine database. The results indicated statistically significant overexpression of HMOX1 in the majority of prostate cancer tissues as compared to normal tissues (Figure 4C; P<0.05). Next, we evaluated the expression of HMOX1 in 10 ADPC and 10 AIPC tissue specimens by IHC staining. The results showed that HMOX1 was more highly expressed in AIPC tissues versus ADPC tissues (Figure 4D; P<0.05).
To further confirm the effect of AIPC-derived exosomes on the expression of HMOX1 in ADPC cells, Western blotting and qRT-PCR were used to determine protein and mRNA levels of HMOX1. The results indicated that HMOX1 protein and mRNA level in LNCAP cells were significantly increased after treatment with PC-3 exosomes (Figure 4E and F; P<0.05).
Activation of HMOX1 Promotes ADPC Cell Progression Resulting in Reduced Androgen Dependence
To investigate the effect of HMOX1 on androgen dependence of ADPC cells, we first used hemin and siRNA to up-regulate and down-regulate, respectively, the expression of HMOX1 in LNCAP cells. The qRT-PCR and Western blotting analysis showed that hemin up-regulated and siRNA down-regulated HMOX1 (Figure 5A and B; P<0.01). MTS assay results indicated that under the normal condition there was no significant difference in cell proliferation between normal LNCAP cells or those treated with hemin (Figure 5C; P>0.05). However, in the absence of androgen up-regulation of HMOX1 expression promoted LNCAP cell proliferation (Figure 5C; P<0.05). On the other hand, when HMOX1 expression was blocked the proliferation of LNCAP cells was inhibited under the normal condition (Figure 5C; P<0.05). In the absence of androgen, no LNCAP cells were able to grow (Figure 5C).
Discussion
Prostate cancer will develop androgen independence following long-term androgen deprivation therapy. The transformation of prostate cancer from androgen-dependent to androgen-independent is a multistep process, and during this process, ADPC cells and AIPC cells may exist at the same time. Hence, cell–cell interactions may contribute to the transition of ADPC cells into AIPC cells. Our data showed that co-culture with PC-3 cells increased the proliferation of LNCAP cells in androgen-deprived medium, and weaken dependence on the AR signaling pathway. However, the mechanism by which PC-3 cells confer castration resistance of LNCAP cells remains unclear.
Exosomes are important mediators of cell–cell interactions and play multiple roles in maintaining normal physiological activities and regulating a variety of pathophysiological processes, including inflammatory, autoimmune and neurodegenerative diseases.17–19 Donor cells can transfer their own specific proteins, lipids or nucleic acids to recipient cells using exosome as a “medium” and thus induce characteristics to the recipient cells such that they are better able to adapt to the changes in the environment.20,21 Accumulating evidence suggests that exosomes play an essential role in the progression of prostate cancer. Hosseini-Beheshti et al reported that prostate cancer-derived exosomes promoted cell proliferation and migration in LNCAP cells.22 Corcoran et al found that the docetaxel-sensitive DU145, 22RV1 and LNCAP parent cells were conferred a significant increase in resistance to docetaxel by transfer of MDR-1/P-gp VIA exosomes.10 However, the function of exosomes in the development of androgen insensitive prostate cancer from androgen-sensitive prostate cancer has not yet been determined.
In this study, we found that the proliferation and tumorigenicity of LNCAP cells were significantly enhanced after exposure to PC-3 exosomes. Under the castration condition, normal LNCAP cells failed to grow, whereas after treatment with PC-3 exosomes LNCAP cells proliferated and formed tumors. We also found that the androgen sensitivity of LNCAP cells decreased significantly in the presence of PC-3 exosomes as compared to LNCAP cells exposed to the own exosomes. These results are consistent with the results of LNCAP/PC-3 cell co-culture experiments and suggest that AIPC exosomes may have the ability to promote the transformation of ADPC cells into androgen-independent cells.
Previous studies have shown that AR expression is associated with the proliferation, migration and invasion of prostate cancer.23,24 Hsieh et al reported that hyper-activation of AR in LNCAP cells resulted in androgen-independent cell proliferation while the parental LNCAP cells only grew in an androgen-dependent manner,25 suggesting AR may play an important role in the androgen-independent growth of prostate cancer cells. However, our study found that the androgen-independent growth of LNCAP cells as a result of stimulation by PC-3 exosomes did not depend on activation of the AR pathway. Hence, we hypothesize that other signaling pathways may be activated by PC-3 exosomes to promote LNCAP cell growth in an androgen-deprived condition. Hence, exploring the mechanisms by which exosomes modulate the androgen sensitivity of recipient cells may provide methods for delaying progression to CRPC.
To further identify the mechanisms of AIPC exosomes-induced castration resistance, Affymetrix microarray technique was used to detect differentially expressed genes of LNCAP cells before and after treatment with PC-3 exosomes. Nine genes with a ≥ 2-fold difference in expression were identified: RARRES3, AKR1C1, SERPINA3, HMOX1, DHRS3, S100P, SAA1, HINT3, and MMP7. The GO analysis revealed that the differentially expressed genes were mainly related to biological processes of inflammation, defense response, and complement binding. Furthermore, KEGG pathway analysis showed that the differentially expressed genes are mainly involved in the regulation and activation of complement, and cancer-related pathways.
HMOX1 is a member of the heat shock protein family (heat shock protein 32) and is a rate-limiting enzyme of heme degradation, which is critical for the inflammatory response.26 HMOX1 had been reported to be highly expressed in various solid tumors, and have an important role in tumor growth.27 Alaoui-Jamali et al reported that HMOX1 expression is higher in CRPC tissues than in ADPC or benign prostate tissue. The study also reported that down-regulation of HMOX1 expression inhibited the activation of the MAPK/p38 signaling pathway, decreased the cell proliferation, survival, and invasion in vitro, and inhibited tumor growth and metastases in vivo.16 Moreover, HMOX1 was found to negatively regulate the transcriptional activity of AR by interfering with STAT3 signaling. Another study showed that induction of HMOX-1 in prostate cancer inhibits AR activation by decreasing PSA promoter activity and mRNA levels.28 The results of the current study confirmed that HMOX1 expression in LNCAP cells was significantly increased after exposure to PC-3 exosomes, demonstrating that PC-3 cells could up-regulate HMOX1 expression in LNCAP cells in a transcellular manner.
To verify the involvement of HMOX1 in androgen resistance, we compared HMOX1 expression in 10 ADPC tissue specimens and 10 AIPC tissue specimens by IHC staining. The overall staining intensity of HMOX1 in AIPC tissues was higher than that in ADPC tissues. By up-regulating the expression of HMOX1, the cell proliferation was significantly enhanced than those of normal LNCAP cells in the absence of androgen; on the contrary, down-regulation of HMOX1 led to a significant decrease of cell viability in LNCAP cells under normal condition. These results taken together, suggest that HMOX1 may be a promising novel therapeutic target for CRPC patients, and additional studies are warranted.
In conclusion, our results indicate that AIPC cells promote the transition of ADPC cells into androgen-independent cells via exosome-mediated activation of HMOX1. Our findings may contribute to the elucidation of the role of the tumor microenvironment in the development of CRPC, and provide a new direction for treatment.
Ethical Approval
All animal experiments were performed by skilled experimenters under the approval of the Medical Ethics committee of Zhujiang Hospital of Southern Medical University, which followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Paraffin-embedded prostate cancer tissues were taken from patients undergoing surgical resection or biopsy at Zhujiang Hospital (Guangzhou, China), and that these experiments were conducted in accordance with the Declaration of Helsinki. And the informed consent was obtained from all patients.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81802516).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Cornford P, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur Urol. 2017;71(4):630–642. doi:10.1016/j.eururo.2016.08.002
3. Mohla S, Stearns V, Sathyamoorthy N, Rosenfeld MG, Nelson P. The biology of hormone refractory breast and prostate cancer: an NCI workshop report. Cancer Biol Ther. 2009;8(21):1975–1985. doi:10.4161/cbt.8.21.9918
4. Lin Y, Fukuchi J, Hiipakka RA, Kokontis JM, Xiang J. Up-regulation of Bcl-2 is required for the progression of prostate cancer cells from an androgen-dependent to an androgen-independent growth stage. Cell Res. 2007;17(6):531–536. doi:10.1038/cr.2007.12
5. Linja MJ, Savinainen KJ, Saramäki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61(9):3550–3555.
6. Golias C, Iliadis I, Peschos D, Charalabopoulos K. Amplification and co-regulators of androgen receptor gene in prostate cancer. Exp Oncol. 2009;31(1):3–8.
7. Ford OR, Gregory CW, Kim D, Smitherman AB, Mohler JL. Androgen receptor gene amplification and protein expression in recurrent prostate cancer. J Urol. 2003;170(5):1817–1821. doi:10.1097/01.ju.0000091873.09677.f4
8. Veldscholte J, Berrevoets CA, Ris-Stalpers C, et al. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J Steroid Biochem Mol Biol. 1992;41(3–8):665–669. doi:10.1016/0960-0760(92)90401-4
9. Liang AL, Du SL, Zhang B, et al. Screening miRNAs associated with resistance gemcitabine from exosomes in A549 lung cancer cells. Cancer Manag Res. 2019;11:6311–6321. doi:10.2147/CMAR.S209149
10. Corcoran C, Rani S, O’Brien K, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7(12):e50999. doi:10.1371/journal.pone.0050999
11. Lundholm M, Schröder M, Nagaeva O, et al. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: mechanism of immune evasion. PLoS One. 2014;9(9):e108925. doi:10.1371/journal.pone.0108925
12. Chowdhury R, Webber JP, Gurney M, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget. 2015;6(2):715–731. doi:10.18632/oncotarget.2711
13. Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi:10.1016/j.eururo.2014.07.035
14. Zhu L, Qu XH, Sun YL, Qian YM, Zhao XH. Novel method for extracting exosomes of hepatocellular carcinoma cells. World J Gastroenterol. 2014;20(21):6651–6657. doi:10.3748/wjg.v20.i21.6651
15. Wei JX, Lv LH, Wan YL, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61(4):1284–1294. doi:10.1002/hep.27660
16. Alaoui-Jamali MA, Bismar TA, Gupta A, et al. A novel experimental heme oxygenase-1-targeted therapy for hormone-refractory prostate cancer. Cancer Res. 2009;69(20):8017–8024. doi:10.1158/0008-5472.CAN-09-0419
17. Conigliaro A, Fontana S, Raimondo S, Alessandro R. Exosomes: nanocarriers of biological messages. Adv Exp Med Biol. 2017;998:23–43.
18. Lopez-Verrilli MA, Picou F, Court FA. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia. 2013;61(11):1795–1806. doi:10.1002/glia.22558
19. Frühbeis C, Fröhlich D, Kuo WP, et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11(7):e1001604. doi:10.1371/journal.pbio.1001604
20. Li W, Zhang X, Wang J, et al. TGFβ1 in fibroblasts-derived exosomes promotes epithelial-mesenchymal transition of ovarian cancer cells. Oncotarget. 2017;8(56):96035–96047. doi:10.18632/oncotarget.21635
21. Schillaci O, Fontana S, Monteleone F, et al. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: their emerging role in tumor heterogeneity. Sci Rep. 2017;7(1):4711. doi:10.1038/s41598-017-05002-y
22. Hosseini-Beheshti E, Choi W, Weiswald LB, et al. Exosomes confer pro-survival signals to alter the phenotype of prostate cells in their surrounding environment. Oncotarget. 2016;7(12):14639–14658. doi:10.18632/oncotarget.7052
23. Brown NE, Paluch AM, Nashu MA, Komurov K, Waltz SE. Tumor cell autonomous RON receptor expression promotes prostate cancer growth under conditions of androgen deprivation. Neoplasia. 2018;20(9):917–929. doi:10.1016/j.neo.2018.07.003
24. He Y, Hooker E, Yu EJ, et al. Androgen signaling is essential for development of prostate cancer initiated from prostatic basal cells. Oncogene. 2019;38(13):2337–2350. doi:10.1038/s41388-018-0583-7
25. Hsieh CL, Cai C, Giwa A, et al. Expression of a hyperactive androgen receptor leads to androgen-independent growth of prostate cancer cells. J Mol Endocrinol. 2008;41(1):13–23. doi:10.1677/JME-07-0158
26. Jozkowicz A, Was H, Dulak J. Heme oxygenase-1 in tumors: is it a false friend? Antioxid Redox Signal. 2007;9(12):2099–2117. doi:10.1089/ars.2007.1659
27. Fang J, Akaike T, Maeda H. Antiapoptotic role of heme oxygenase (HO) and the potential of HO as a target in anticancer treatment. Apoptosis. 2004;9(1):27–35. doi:10.1023/B:APPT.0000012119.83734.4e
28. Elguero B, Gueron G, Giudice JF, et al. Unveiling the association of STAT3 and HO-1 in prostate cancer: role beyond heme degradation. Neoplasia. 2012;14(11):1043–1056. doi:10.1593/neo.121358
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.