Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
Exosomes from β-Cells Promote Differentiation of Induced Pluripotent Stem Cells into Insulin-Producing Cells Through microRNA-Dependent Mechanisms
Authors Guo Q, Lu Y, Huang Y, Guo Y, Zhu S, Zhang Q, Zhu D, Wang Z, Luo J
Received 6 October 2021
Accepted for publication 1 December 2021
Published 11 December 2021 Volume 2021:14 Pages 4767—4782
DOI https://doi.org/10.2147/DMSO.S342647
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ming-Hui Zou
Qingsong Guo,1,2,* Yuhua Lu,1,* Yan Huang,1,2 Yibing Guo,2 Shajun Zhu,1,2 Qiuqiang Zhang,1 Donghui Zhu,3 Zhiwei Wang,1 Jia Luo4
1Department of Hepatobiliary and Pancreatic Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China; 2Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China; 3Nantong University Medical School, Nantong, 226001, People’s Republic of China; 4Department of Pharmacy, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jia Luo
Department of Pharmacy, Affiliated Hospital of Nantong University, Xi Si Road, Nantong, 226001, People’s Republic of China
Email [email protected]
Zhiwei Wang
Department of Hepatobiliary and Pancreatic Surgery, Affiliated Hospital of Nantong University, Xi Si Road, Nantong, 226001, People’s Republic of China
Email [email protected]
Objective: Exosomes have emerged as potential tools for the differentiation of induced pluripotent stem cells (iPSCs) into insulin-producing cells (IPCs). Exosomal microRNAs are receiving increasing attention in this process. Here, we aimed at investigating the role of exosomes derived from a murine pancreatic β-cell line and identifying signature exosomal miRNAs on iPSCs differentiation.
Methods: Exosomes were isolated from MIN6 cells and identified with TEM, NTA and Western blot. PKH67 tracer and transwell assay were used to confirm exosome delivery into iPSCs. qRT-PCR was applied to detect key pancreatic transcription gene expression and exosome-derived miRNA expression. Insulin secretion was determined using FCM and immunofluorescence. The specific exosomal miRNAs were determined via RNA-interference of Ago2. The therapeutic effect of 21 day-exosome-induced IPCs was validated in T1D mice induced by STZ.
Results: iPSCs cultured in medium containing exosomes showed sustained higher expression of MAFA, Insulin1, Insulin2, Isl1, Neuroud1, Nkx6.1 and NGN3 compared to control iPSCs. In FCM analysis, approximately 52.7% of the differentiated cells displayed insulin expression at the middle stage. Consistent with the gene expression data, immunofluorescence assays showed that Nkx6.1 and insulin expression in iPSCs were significantly upregulated. Intriguingly, the expression of pancreatic markers and insulin was significantly decreased in iPSCs cultured with siAgo2 exosomes. Transplantation of 21 day-induced IPCs intoT1D mice efficiently enhanced glucose tolerance and partially controlled hyperglycemia. The therapeutic effect was significantly attenuated in T1D mice that received iPSCs cultured with siAgo2 exosomes. Of the seven exosomal microRNAs selected for validation, miR-706, miR-709, miR-466c-5p, and miR-423-5p showed dynamic expression during 21 days in culture.
Conclusion: These data indicate that differentiation of exosome-induced iPSCs into functional cells is crucially dependent on the specific miRNAs encased within exosomes, whose functional analysis is likely to provide insight into novel regulatory mechanisms governing iPSCs differentiation into IPCs.
Keywords: diabetes, exosomes, iPSCs, differentiation, miRNA, insulin
Introduction
Diabetes mellitus (DM) is one of the most common chronic diseases worldwide and The International Diabetes Federation (IDF) estimated the global population with diabetes to be an exponential growth from 463 million in 2019 to 700 million in 2045.1,2 It is primarily characterized by dysfunctional pancreatic islets or the loss of insulin-producing β-cells.3–6 Almost all current treatments rely on daily insulin injections or the transplantation of cadaveric islets. Replenishment or regeneration of lost pancreatic β-cells by alternative cells to restore β-cell function is an ideal long-term clinical solution. Although pancreatic and islet transplantation has provided an opportunity to cure diabetes,7,8 the approach is still limited by the shortage of suitable donors, poor function and viability of pancreatic islets isolated from donors, and immune rejection.9,10 Stem cell-derived β-cell therapy has been proposed as a convenient solution to the existing shortcomings.
Induced pluripotent stem cells (iPSCs) can be differentiated into insulin-producing cells (IPCs) by sequential application of exogenous molecules in vitro and used as an alternative source of β-cells for therapy.3 However, some DNA-based reprogramming techniques have inherent biosafety concerns, such as insertional mutagenesis or tumorigenesis.11–13 Additionally, the functionality, differentiation, and maturation of iPSCs using these methods is still low.14 To this end, a strategy for efficient differentiation of iPSCs into functional IPCs is urgently required and will fuel future research in β-cell therapy.
Exosomes are microvesicles (30–150 nm in diameter) that originate from late endosomes, have a functional membranous structure and can be secreted into the extracellular space, transferring bioactive components to other cells.15 Intercellular delivery of exosomes plays an important role in modifying or reprogramming the phenotype of recipient cells.16 Pancreatic islets and various β-cell lines have been reported to release exosomes carrying noncoding RNAs and proteins.17–19 Given the advantages of higher stability, biosafety, easy accessibility, and controllable dosage, exosomes are promising tools that could promote stem cell differentiation.
Specific miRNAs are selectively packaged into exosomes and transferred to recipient cells, thus regulating post-transcriptional gene expression that affects stem cell differentiation.20 miRNAs directly or indirectly target islet cell-specific genes, which results in translational repression or degradation of specific mRNAs, thus regulating islet cell differentiation and maturation.21–24 Ago2 is integral to miRNA function as well as the packaging of miRNAs into exosomes.25–27 It has been reported that the function of exosomal miRNA is dependent on Ago2 derived from the donor cells instead of recipient cells. Therefore, knockdown of Ago2 in the host cells allows the isolation and testing of miRNA-depleted exosomes.28,29 Since miRNAs were described as a major noncoding RNA species shuttled through exosomal transfer and activated signaling pathways at the recipient cells or tissues, here, we focused on their composition in exosomes released from MIN6 cells. The identification of the candidate miRNAs would help us better understand the molecular events involved in stem cell differentiation and provide insight into novel regulatory mechanisms governing iPSCs differentiation into IPCs.
Materials and Methods
Cell Culture
The MIN6 cell line was purchased from the American Type Culture Collection and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) exosome-depleted fetal bovine serum (FBS) (Gibco, USA), 100 U/mL penicillin, 100 µg/mL streptomycin (Thermo-Fisher) and cultured at 37°C in a 5% CO2 incubator. Cultivated cells were passaged at 90% confluency.
The iPSCs induced via a three-step protocol in our laboratory30 were maintained in DMEM/F12 conditioned medium with 10% (v/v) FBS, 100 U/mL penicillin and 100 µg/mL streptomycin at the multi-lineage pluripotent cell (MPC) stage. MPCs medium was replaced with differentiation medium containing complete knockout DMEM; 10% exosome-depleted FBS; MIN6-derived exosomes; and 1×penicillin-streptomycin. The culture medium was changed twice a day.
Argonaute-2 (Ago2) Knockdown in MIN6 Cells
MIN6 cells were transfected with Ago2 siRNA (siAgo2) (Ruibo Biotech, China) using Lipofectamine 3000 (Thermo-Fisher) according to the manufacturer's protocol. Briefly, MIN6 cells at 80% confluence were incubated with lipoplexes formed with Lipofectamine 3000 and siAgo2 or a scrambled control siRNA (siNC) for 48 h. Quantitative polymerase chain reaction (qPCR) and Western blot were performed to verify the efficiency of knockdown.
Isolation of Exosomes
Passage 3 MIN6 cells grown to 80% confluence in T75 flask (transfected with siAgo2, siNC or nontransfected) were washed 3 times with phosphate-buffered saline (PBS) and then incubated in exosome-depleted FBS medium. The supernatant were collected every other day for exosome isolation. Briefly, the supernatant was centrifuged at 2000 g to remove dead cells for 10 min and again at 11,000 g for 30 min to remove cell debris at 4°C. The supernatant was filtered through a 0.22 μm filter (Millipore-sigma). Ultracentrifugation was performed at 150,000 g for 2 h at 4°C followed by an additional wash with PBS at 150,000 g for 90 minutes. The pellets from every 50 mL supernatant were collected, resuspended in 100 μL PBS, checked for mycoplasma contamination and stored at −80°C for further experiments. Every 100 μL exosomes in PBS were diluted in high glucose-DMEM (H-DMEM, Gibco) medium to a final 10 mL volume with exosomes or siAgo2-exosoems.
Identification and Quantification of Exosomes
Transmission electron microscopy (TEM) was performed to observe the morphology of the exosomes. In brief, 10 μL of the exosome suspension was placed on formvar/carbon-coated nickel TEM grids and incubated for 30 min. The grids were then washed, dried, and imaged using an H-7650 transmission electron microscope (HITACHI, Japan) to characterize the morphology.
Nanoparticle tracking analysis (NTA) was performed using LM10 Nanosight (Malvern) equipped with a 405 nm laser following the manufacturer’s instructions. Exosomes were diluted with PBS (1:100) before analysis. NTA post-acquisition settings were optimized and kept constant between samples, three separate fields of view were captured for each sample for 30 s at room temperature. The Nanosight NTA 3.1 software was used to capture and analyze the particles.
Exosome Labeling and Uptake by iPSCs
Exosomes were labeled using the PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich) according to the manufacturer’s protocol. Specifically, 100 μL exosomes were suspended in 250 μL Diluent C, to which 4μL PKH67 dye was added and incubated with the exosome solution for 4 min. 1 mL of 0.5% BSA was added to bind excess dye. The labeled exosomes were ultracentrifuged at 100,000 g for 70 min, after which the pellets were washed with PBS and ultracentrifuged again. The pellets were diluted in 100 μL of PBS and used for uptake experiments. 10 μL labeled exosomes were incubated with iPSCs for 24 h, the medium was discarded and the iPSCs were washed three times with PBS. The fluorescence signals among the iPSCs were detected by fluorescence microscopy (Olympus BX41, Japan) to determine whether fusion had occurred between the added exosomes and iPSCs.
Transwell (Corning) inserts were used to co-culture MIN6 cells with iPSCs. MIN6 cells were seeded with the density of 5×104 cells/mL in the top chamber of the inserts and placed into 24-well culture plates seeded with the density of 5×104 cells/mL iPSCs. MIN6 cells were transfected with the positive control Cy3-labelled siRNA (Ruibo Biotech, China). The red fluorescence in MIN6 cells was observed after 24 h and in the iPSCs after 48 h under fluorescence microscopy.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was extracted using the Trizol reagent and the first-strand cDNA synthesis for Ago2 and other genes was performed using the RevertAid First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s instructions. The relative expression level of each mRNA was calculated by the 2−ΔΔCt method, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the control gene (U6 was used as the control gene for miRNAs). Each experiment was independently repeated three times and each sample was set with three duplicate wells. Primer (Ruibo Biotech, China) sequences are shown in Table 1.
 |
Table 1 Polymerase Chain Reaction Primer Sequences |
Immunofluorescence
Cells grown to 80% confluency on glass coverslips were washed with pre-cooled PBS and fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.5% (v/v) Triton X-100. The cells were incubated with 5% bovine serum albumin (BSA) for 1 h and then with specific primary antibodies at 4°C overnight. The cells were stained with fluorescent secondary antibodies and Hoechst (Thermo Fisher Scientific). The primary antibodies used in this study are as follows: anti-insulin antibody (Ab) (1:500, Abcam) (Cat. ab181547), anti-Nkx6.1 rabbit monoclonal Ab (1:200, Cell Signaling Technology) (Cat. #54551S), and anti-glucagon Ab (1:1000, Abcam) (Cat. ab92571). Secondary antibodies included donkey anti-rabbit Ab (1:1000, Alexa Fluor® 488, Abcam) (Cat. ab150133) and (1:1000, Alexa Fluor® 647, Abcam) (Cat. ab150187).
Flow Cytometry
We performed FCM to identify the insulin-positive cell population. About 1×106 differentiated iPSCs on day 7 and day 14 were digested with 0.25% trypsin, washed, and resuspended as single cells in Reagent 1: Fixation (Beckman Coulter). The cells were then incubated in Reagent 2: Permeabilization (Beckman Coulter) and washed with PBS. Next, the cells were resuspended in PBS containing the primary antibody, incubated for 30 min, washed, and analyzed with the BD FACSCalibur system (BD Biosciences). The primary antibody was APC-conjugated anti-h/b/m insulin rat IgG2A (R&D Systems). The isotype antibody was APC-conjugated rat IgG2A control (BD Biosciences).
Western Blot
BCA Protein Assay Kit (Sangon Biotech, China) was used for the protein quantification. 40μg exosomal proteins and cell lysates were separated by 10% sodium SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, USA), blocked with 5% skim milk, and probed with primary antibodies against Alix (1:1000, Abcam, USA) (Cat. ab275377), TSG101 (1:1000, Abcam) (Cat. ab125011), Ago2 (1:1000, Abcam) (Cat.ab186733) at 4°C overnight. The membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology) after washing with Tris-buffer saline Tween-20 (TBST). The blots were then developed using an ECL kit (Thermo Fisher Scientific). Densitometric analysis of the bands was performed using the Multi Gauge software V3.0 from Fuji Film (Minato, Tokyo, Japan).
Animals
C57BL/6J male mice aged 8 weeks were obtained from the Laboratory Animal center of the Nantong University. For the experimental type-I diabetic hyperglycemia model, mice were administered a single intraperitoneal dose of 200 mg/kg STZ (Sigma, USA), and non-fasting blood glucose levels in the tail vein were monitored continuously by a glucose meter (Roche, Shanghai, China). Two weeks later, 24 mice with high blood glucose (>14.4 mg/mL) were randomly divided into three groups, the STZ + iPSCs, STZ+ exosome-induced iPSCs and STZ + siAgo2-exosome-induced iPSCs group (n = 8 for each group). iPSCs (1×107) cultured in the medium containing exosomes over the 21-day period were implanted into the kidney subcapsular space and the blood glucose was measured before and after transplantation. On post-transplant days 7, 14, and 21, the mice were fasted for 8 h and then subjected to an intraperitoneal glucose tolerance test (IPGTT) by administering an intraperitoneal injection of 10% glucose solution (2 mg/kg). All animal experiments were performed according to the Institutional Animal Care guidelines and were approved by the Animal Ethics Committee of Nantong University.
Statistical Analysis
Data were analyzed using GraphPad Prism Ver. 10.0. One-way ANOVA followed by Tukey’s or Dunnett’s post hoc tests or two-way ANOVA was performed for multiple group comparisons as appropriate, and data are presented as the mean ± standard deviation (SD). A p < 0.05 was considered statistically significant.
Results
Isolation and Characterization of Exosomes Released by MIN6 Cells
We first wanted to profile the exosomes produced by β-cells. Alix and TSG101, typical exosomal markers, were highly expressed in the exosomes isolated from MIN6 cells (Figure 1A). TEM was performed on exosomes re-suspended in Tris-buffer saline (TBS) and revealed a diverse population of vesicles (Figure 1B). Using NTA, the majority of the purified microvesicles were found to be 60–200 nm in diameter, with an average size of 140.3 nm, and the subcellular structures exhibited a typical double-layer membrane structure (Figure 1C).
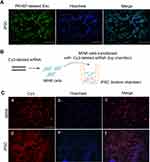
MIN6-Derived Exosomes Successfully Integrated and Delivered Cargos to iPSCs
To investigate exosome uptake by iPSCs, purified exosomes labeled with the green fluorescent marker PKH67 were incubated with iPSCs. Almost all of iPSCs exhibited green fluorescence. PKH67-labeled exosomes located in the cytoplasm, indicating that exosomes were fused into iPSCs when analyzed at 24 h (Figure 2A).
Next, a transwell co-culture assay system with 0.4-μm porous membrane was employed to study the crosstalk between MIN6 and iPSCs. Fluorescence imaging of the filter demonstrated that MIN6 cells were successfully transfected with Cy3-labeled siRNA. As MIN6 cells were unable to migrate through the 0.4 μm porous membrane filter, the Cy3-labeled components must have been released from the MIN6 cells and subsequently taken up by the iPSCs, as red fluorescence was observed in the iPSCs 48 h later (Figure 2B and C).
The concordance of the two experiments confirmed that MIN6 cell-derived exosomes successfully integrated and delivered their cargos to iPSCs.
Expression of Pancreatic Marker Genes in iPSCs Following Exposure to Exosomes at an Early Stage of Differentiation
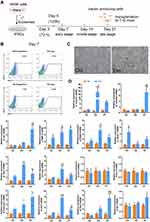
A flow chart illustrating the detailed differentiation process is depicted in Figure 3A.
After seven days of treatment with the β-cell exosomes, FCM was performed to determine the efficiency of iPSC differentiation to form insulin-positive (insulin+) cells. Insulin+ cells accounted for 22.3% of the final population in iPSCs treated with exosomes and only 11.9% of the final population in control cells without exposure to exosomes (Figure 3B), representing a 2-fold increase in insulin+ cells. The iPSCs showed a cluster-like morphology and several cell aggregates were observed (Figure 3C). Exosomes had no effect on cell viability detected by CCK8 assay (Beyotime, China) (data not shown).
We next analyzed the expression of endocrine markers (insulin1, insulin2, GCG, SST, GCK, Isl1, Glut2) and β-cell specific genes (NGN3, Neuroud1 and MAFA), as well as genes expressed during specific stages of pancreatic development (Nkx6.1, Pdx1, Foxa2). Our results confirmed that expression of the Mafa A and Ngn3, was significantly higher in exosome-induced iPSCs than in the control cells. The expression of Foxa2 and Pdx1, critically involved in early pancreatic development, was also analyzed. Indeed, the expression of Foxa2 and Pdx1 increased on day 5 but tended to be lower on day 7. The mRNA expression of the hormone markers, Ins1, Ins2, and GCG was significantly higher in exosome-induced iPSCs than that in control cells. In addition, Nkx6.1, Neuroud1, SST, GCK and Isl1 were significantly upregulated in the early stages of induction compared with their expression in iPSCs without exosomal induction, thus demonstrating the endocrine pancreatic lineage commitment of these cells during the differentiation process. Unexpectedly, the expression of Glut2 was inconsistent with that reported in previous studies, demonstrating very low expression in the induced cells (data not shown). Together, the transcriptional changes confirmed that β-cell exosomes induced differentiation of iPSCs into IPCs, and the process specifically mimics endocrine pancreas organogenesis. The canonical embryonic/iPSC marker Oct4 and endoderm marker Sox17 were detected in the iPSC clusters. However, there was no significant change in the expression of these two genes from day 3 to day 7 (Figure 3D).
Expression of Nkx6.1 and Insulin Protein in Exosome-Induced iPSCs at an Early Stage of Differentiation
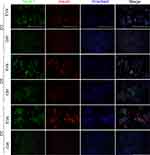
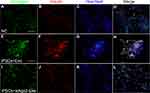
To further verify whether exosomes promoted the expression of specific proteins in iPSCs, immunofluorescence imaging for Nkx6.1 and insulin was performed. On day 3, almost no expression was detected in both exosome-induced cells and in control cells. On day 5, both Nkx6.1 and insulin expression were observed in exosome-induced iPSCs. On day 7, the expression of insulin increased significantly in exosome-induced iPSCs compared with that on day 5 (Figure 4).
Ago2 Inactivation in MIN6 Cells Lowers Gene Expression Profile Associated with Differentiation
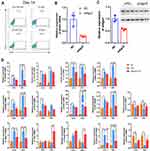
On day 14, the distribution of insulin+ cells was analyzed using FCM. The results confirmed that the proportion of insulin+ cells increased to 52.7% in the presence of exosomes compared with 22.4% in the control group. Compared with that on day 7, the proportion of insulin+ cells on day 14 in the presence of exosomes was 2-fold higher (Figure 5A), suggesting that exosome-derived molecules could stably induce differentiation and insulin expression in iPSCs.
To test whether the increase in gene expression was related to exosomal miRNAs, MIN6 cells were transfected with siRNA targeting Ago2 (siAgo2) to reduce miRNA synthesis prior to the preparation of exosomes (siAgo2-exosomes). qPCR revealed a 70% decrease in Ago2 mRNA expression and approximately 50% decrease in Ago2 protein levels (Figure 5B and C).
Expression of most endocrine and β-cell-specific genes, as well as genes enriched during the early stages, was lowered upon exposure to siAgo2-exosomes (Figure 5D). Isl1, Ins2, Isl1, MafaA, GCG, GCK, Ngn3, and SST expression was not upregulated on day 14 and day 21 (GCG was only upregulated on day 14) compared with iPSCs treated with control exosomes. Nkx6.1 and Neuroud1 expression was synchronous with MafaA expression in exosome-induced iPSCs. Importantly, we found that expression of Foxa2 and Pdx1 was decreased, while increased in siAgo2-exosome-induced iPSCs at the late stages. In addition, the typical embryonic stem cell marker Oct4 and endoderm marker Sox17 were significantly downregulated in iPSC clusters induced with siAgo2-exosomes on both day 14 and day 21. siRNA had no measurable effect on cell viability using CCK8 (Beyotime, China) (data not shown). Taken together, these results suggested that exosomes stably induce iPSCs differentiation in vitro and this process is mediated by exosomal miRNAs.
Glucagon and Insulin Expression at the Late Stage of Differentiation
Immunofluorescence analysis of insulin and glucagon (a marker for α-cells which is another islet cell type) expression was performed to verify the efficiency of late stage differentiation in vitro. We found that both insulin and glucagon displayed weak positive signals in control iPSCs. Additionally, there was no obvious morphological change and the cells maintained the original pleomorphic state of the iPSCs. Surprisingly, both insulin and glucagon were highly expressed in exosome-induced iPSCs. These cells tended to aggregate, grow in clusters, and became larger and rounder compared to undifferentiated iPSCs. Expression of the two hormones was diminished in siAgo2-exosome-induced iPSCs, and a large proportion of cells remain in pleomorphic state, unable to fully differentiate. These results indicated that knockdown of Ago2 prevented exosome-mediated differentiation of iPSCs through complex miRNA-dependent pathway (Figure 6).
Differential Expression of microRNAs in MIN6 Cells, Exosomes and iPSCs
Previous high throughput sequencing (GSE159029) and microarray profiling (232/650 miRNAs) reported 24 reduplicated MIN6-derived miRNAs, among which seven microRNAs showed a high degree of fold-change between MIN6-derived exosome versus MIN6 cells (Figure 7A). The other 17 microRNAs showed low expression in exosomes or had an almost equal representation in MIN6 parental cells and exosomes. Seven selected miRNAs were confirmed in MIN6 cells, MIN6 exosomes, siAgo2-MIN6 cells, siAgo2-MIN6 exosomes (Figure 7B). To validate the differential expression in iPSCs, we analyzed their expression by qPCR at three stages (miRNA information is listed in Table 2). The data demonstrated that miR-706 was gradually expressed from day 7 and increased up to day 21 in exosome-induced iPSCs, while in siAgo2-exosome induced iPSCs, the expression increased on day 7 and then decreased during the late stage. The miR-709 was significantly upregulated on day 14 and peaked on day 21 in exosome-induced iPSCs compared to the control cells. The miR-466c-5p demonstrated an expression pattern similar to that of miR-709. However, miR-423-5p showed maximum upregulation in the early stage, and gradually decreased during the late stage. The miR-1187 expression in siAgo2-exosome induced iPSCs was significantly reduced on day 14 and day 21. miR-1895 and miR-671-5p showed no obvious changes during iPSC differentiation (Figure 7C). Thus, the expression of various miRNAs dynamically change during differentiation and this process was influenced by exosomes.
 |
Table 2 List of miRNAs Detected During Differentiation Stages in the Experiment |
Differentiated IPCs Improve Glycemic Control in a Murine Model of Experimental Diabetic Hyperglycemia
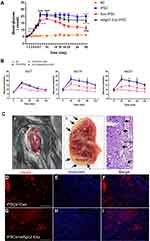
Using a murine model of experimental diabetic hyperglycemia, the functionality of the differentiated IPCs was explored in vivo. Diabetic mice were infused with control iPSCs, exosome-induced iPSCs, or siAgo2-exosome-induced iPSCs and the non-fasting blood glucose was monitored. The results showed that the non-fasting blood glucose concentration in control non-diabetic mice remained stable and normal, while blood glucose concentration in the STZ + iPSCs group, STZ + exosome-induced iPSCs group, and STZ + siAgo2-exosome-induced iPSCs group showed much higher blood glucose levels (Figure 8A). On day 7 post-transplantation, blood glucose concentration in both the STZ + exosome-induced iPSCs group and STZ + siAgo2-exosome-induced iPSCs group showed a downward trend. This reduction could be stably maintained for 14 days after transplantation in STZ + exosome-induced iPSCs animals, implying that the transplanted cells effectively controlled blood glucose levels. Although the concentration was not lowered to normal levels as reported, we found a 50% decrease in the blood glucose concentration (10 mmol/L) of STZ + exosome-induced iPSCs group compared to the NC group. Meanwhile, STZ-treated control mice exhibited progressively impaired glucose tolerance on day 7, 14, and 21. The STZ+exosome-induced iPSCs group show much better glycemic control compared to the two other groups on day 14 and 21 (Figure 8B).
Immunofluorescence analysis of frozen kidney tissues revealed that insulin expression in the STZ + exosome-induced iPSCs group was significantly higher than that in the STZ + siAgo2-exosome-induced iPSCs group (Figure 8C), implying that the siAgo2-exosome-induced iPSC graft was functionally deficient. This observation further supported the assertion that knockdown of Ago2 significantly attenuated exosomal miRNA function and impaired iPSC differentiation.
Discussion
Previous studies have demonstrated the application of iPSC-based differentiation protocols for β-like cell regeneration.31–33 The application of iPSCs as islet-derived cells is not limited in ethics, and iPSCs from individual specific sources will not cause immune rejection. Based on the advantages mentioned above, iPSCs can become ideal seed cells for the transplantation of diabetes treatment. However, the differentiation efficiency of iPSCs into functional IPCs is hampered by several technical and biological limitations, which may result in sub-optimal differentiation and immature β-cells. Given their role as important epigenetic regulators, miRNAs reportedly influence stem cell differentiation. Although miRNAs packaged by MIN6 cells into exosomes have been profiled in recent years,34,35 very few studies have specifically elucidated the molecular role of exosomal miRNAs in iPSC differentiation.
In the present study, we found that during the early phase of differentiation, expression of genes related to endocrine pancreatic development, such as mature endocrine cell-specific genes Insulin1, Insulin2, GCG, and SST were upregulated in iPSCs cultured with exosomes. In addition, β-cell specific genes Mafa A, Neuroud1 and NGN3, the transcription factors Nkx6.1, Foxa2 and Pdx1 were also upregulated in these cells. Among these genes, we found that expression of Nkx6.1 and Mafa A show the most significant upregulation in iPSCs cultured with exosomes compared to iPSCs cultured without exosomes. Moreover, the expression of GCK, part of the glucose-responsive machinery, was stably upregulated in iPSCs cultured with exosomes throughout the 21 days in culture. However, the stem cell-specific genes Oct4 and Sox17 showed no significant change during the early stage of differentiation but decreased significantly in the late stage. The population of insulin+ cells accounted for more than half of the total of cells at day 14 and cell viability was not affected by the exosomes stimulation. Since a previous study reported that insulin+ cells co-expressing Nkx6.1 can differentiate into functional β-cells following in vivo transplantation,36,37 we also analyzed the expression of insulin and Nkx6.1 in iPSCs. We found that both insulin and Nkx6.1 remarkably upregulated on day 7, by contrast, there was hardly any expression of insulin and Nkx6.1 in iPSCs cultured without exosomes. These results further confirmed the feasibility to obtain IPCs using exosomes as xeno-free media in the 2-D culture, avoiding viral transfections and making iPSCs more suitable for clinical trials.
Interestingly, siAgo2-exosomes from MIN6 cells showed poor induction of pancreatic gene expression in iPSCs. Further support came from our studies on day 21. In addition to a cell-like structure and high expression of insulin, glucagon (a marker for α-cells, another islet cell type) was also expressed in the cells. The result is inconsistent with the results of a previous study, which believed β cells could not directly induce glucagon production in the process.35 Following transplantation of differentiated IPCs, we found that the graft contributed to blood glucose control in diabetic mice, improved the fasting blood glucose levels, and glucose tolerance. The hyperglycemia in mice was mitigated, implying the transplanted IPCs were functional in a physiological milieu, although blood glucose could not be brought down to normal levels. Histopathological observation showed that IPCs transplanted under the left renal capsule of mice were still alive and still expressed insulin. Immunofluorescence analysis of insulin expression in the transplanted graft revealed that Ago2 knockdown impaired insulin expression, indicating that the essential driving force for differentiation was largely dependent on exosomal miRNAs from the donor cells.
Two databases containing MIN6 exosomal miRNA-seq were analyzed and 24 overlapping miRNAs were selected.35,38 Among them, six microRNAs showed high differential expression between the exosomes and the parental cells, indicating that specific miRNAs were preferentially packaging into exosomes while others may be selectively retained in the parent MIN6 cells. Selective accumulation of specific miRNAs in exosomes released by other cell types have been reported before.39,40 Several miRNAs including, miR-709, miR-706, and miR-466c-5p were upregulated gradually from day 7 to day 21. The expression pattern of miR709 was in line with previous studies and it has been reported to inhibit adipocyte differentiation by targeting GSK3β, and subsequently activating Wnt/β-catenin signaling.41 On the contrary, miR-423-5p, reported to be associated with diabetes,42–44 was gradually downregulated over the 21 days in culture, which further underlined the fact that exosomal-miRNA activity may be required within a narrow time window to regulate iPSC differentiation. However, miR-671-5p and miR-1187 did not show any significant change with exosomal-induction. Based on our data, we found that four exosomal microRNAs (miR-706, miR-709, miR-466c-5p, and miR-423-5p) actively modulate iPSC differentiation. Therefore, it is imperative we focus on these microRNAs in the future for stem cell research in diabetes.
Our findings convincingly confirmed that iPSC differentiation into functional IPCs through the delivery of exosomes was effective and feasible, although the present study was unable to delineate the exact mechanisms of miRNA-gene interaction or identify dominant miRNAs involved in the differentiation process. The population of insulin+ cells induced with exosomes in vitro was 52.7% in the middle of the differentiation process, which was higher than our previous efforts.30 Even so, hyperglycemia in mice was controlled only partially, suggesting that iPSC-based insulin expression was still insufficient to completely regulate blood glucose. Recently, it was reported that IPCs derived from iPSCs in 2-D culture expressed lower levels of mature pancreatic markers such as insulin and C-peptide than did those in 3-D culture.45 Acellular pancreatic scaffold has been applied to improve the yield of functional IPCs. This indicates that the establishment and maintenance of a full maturity and functional organoid still present unique challenges when grown ex vivo. From a more comprehensive perspective, acinar (elastase-specific) and ductal (cytokeratin-specific) in the exosome-treated iPSCs should also be detected during the course of differentiation and maturation in vivo.
Although β-cell differentiation is a complex and highly regulated process, our findings provide crucial insights into the role of exosomal miRNAs in this network. Further studies are required for a more detailed and systematic assessment of exosomal miRNAs, both in vitro and in vivo.
Conclusion
In summary, our study delves into the role of β-cell derived exosomes in the differentiation of iPSCs into IPCs and specific exosomal miRNAs which participated in this process. We demonstrated that these cells were capable of regulating blood glucose in a murine diabetes model and Ago2-knockdown in the parent cells attenuated iPSC differentiation. We hope that our study may lay a foundation for future studies on the exosomal microRNAs regulatory network in stem cell-differentiation and provide a potential therapeutic strategy for diabetic treatment.
Acknowledgments
This work was supported by Project funded by China Postdoctoral Science Foundation (2019M661904) and Postdoctoral program funded by Jiangsu province (2019Z154).
Disclosure
The authors declare no potential conflicts of interest.
References
1. Sinclair A, Saeedi P, Kaundal A, et al. Diabetes and global ageing among 65-99-year-old adults: findings from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2020;162:108078. doi:10.1016/j.diabres.2020.108078
2. He X, Zhang Y, Zhou Y, et al. Direct medical costs of incident complications in patients newly diagnosed with type 2 diabetes in China. Diabetes Ther. 2021;12(1):275–288. doi:10.1007/s13300-020-00967-y
3. Rezania A, Bruin J, Arora P, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. doi:10.1038/nbt.3033
4. Maxwell K, Millman J. Applications of iPSC-derived beta cells from patients with diabetes. Cell Rep Med. 2021;2(4):100238. doi:10.1016/j.xcrm.2021.100238
5. Agrawal A, Narayan G, Gogoi R, et al. Recent advances in the generation of β-cells from induced pluripotent stem cells as a potential cure for diabetes mellitus. Adv Exp Med Biol. 2021. doi:10.1007/5584_2021_653
6. Lu J, Xia Q, Zhou Q. How to make insulin-producing pancreatic β cells for diabetes treatment. Sci China Life Sci. 2017;60(3):239–248. doi:10.1007/s11427-016-0211-3
7. Bellin M, Barton F, Heitman A, et al. Potent induction immunotherapy promotes long-term insulin Independence after islet transplantation in type 1 diabetes. Am J Transplant. 2012;12(6):1576–1583. doi:10.1111/j.1600-6143.2011.03977.x
8. Shapiro A, Lakey J, Ryan E, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343(4):230–238. doi:10.1056/NEJM200007273430401
9. Croon A, Karlsson R, Bergström C, et al. Lack of donors limits the use of islet transplantation as treatment for diabetes. Transplant Proc. 2003;35(2):764.
10. Williams M, Joglekar M, Satoor S, et al. Epigenetic and transcriptome profiling identifies a population of visceral adipose-derived progenitor cells with the potential to differentiate into an endocrine pancreatic lineage. Cell Transplant. 2019;28(1):89–104. doi:10.1177/0963689718808472
11. Haridhasapavalan K, Borgohain M, Dey C, et al. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene. 2019;686:146–159. doi:10.1016/j.gene.2018.11.069
12. Borgohain M, Haridhasapavalan K, Dey C, et al. An insight into DNA-free reprogramming approaches to generate integration-free induced pluripotent stem cells for prospective biomedical applications. Stem Cell Rev Rep. 2019;15(2):286–313. doi:10.1007/s12015-018-9861-6
13. Dey C, Raina K, Haridhasapavalan KK, et al. An overview of reprogramming approaches to derive integration-free induced pluripotent stem cells for prospective biomedical applications. Recent Adv iPSC Technol. 2021;231–287. doi:10.1016/B978-0-12-822231-7.00011-4
14. Zhou Q, Melton D. Pancreas regeneration. Nature. 2018;557(7705):351–358. doi:10.1038/s41586-018-0088-0
15. Katsuda T, Ochiya T. Molecular signatures of mesenchymal stem cell-derived extracellular vesicle-mediated tissue repair. Stem Cell Res Ther. 2015;6:212. doi:10.1186/s13287-015-0214-y
16. Moradi S, Asgari S, Baharvand H. Concise review: harmonies played by microRNAs in cell fate reprogramming. Stem Cells. 2014;32(1):3–15. doi:10.1002/stem.1576
17. Bashratyan R, Sheng H, Regn D, et al. Insulinoma-released exosomes activate autoreactive marginal zone-like B cells that expand endogenously in prediabetic NOD mice. Eur J Immunol. 2013;43(10):2588–2597. doi:10.1002/eji.201343376
18. Castaño C, Novials A, Párrizas M. Exosomes and diabetes. Diabetes Metab Res Rev. 2019;35(3):e3107. doi:10.1002/dmrr.3107
19. Pardo F, Villalobos-Labra R, Sobrevia B, et al. Extracellular vesicles in obesity and diabetes mellitus. Mol Aspects Med. 2018;60:81–91. doi:10.1016/j.mam.2017.11.010
20. Li J, Tong C, Sung T, et al. CMEP: a database for circulating microRNA expression profiling. Bioinformatics. 2019;35(17):3127–3132. doi:10.1093/bioinformatics/btz042
21. LaPierre M, Stoffel M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol Metab. 2017;6(9):1010–1023. doi:10.1016/j.molmet.2017.06.020
22. Zhang A, Li D, Liu Y, et al. Islet β cell: an endocrine cell secreting miRNAs. Biochem Biophys Res Commun. 2018;495(2):1648–1654. doi:10.1016/j.bbrc.2017.12.028
23. Eliasson L. The small RNA miR-375 - A pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Mol Cell Endocrinol. 2017;456:95–101. doi:10.1016/j.mce.2017.02.043
24. Kaviani M, Azarpira N, Karimi M, et al. The role of microRNAs in islet β-cell development. Cell Biol Int. 2016;40(12):1248–1255. doi:10.1002/cbin.10691
25. Ha M, Kim V. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi:10.1038/nrm3838
26. Tattikota S, Rathjen T, McAnulty S, et al. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metab. 2014;19(1):122–134. doi:10.1016/j.cmet.2013.11.015
27. Guduric-Fuchs J, O’Connor A, Camp B, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012;13:357. doi:10.1186/1471-2164-13-357
28. Zhang Y, Chopp M, Liu X, et al. Exosomes derived from mesenchymal stromal cells promote axonal growth of cortical neurons. Mol Neurobiol. 2017;54(4):2659–2673. doi:10.1007/s12035-016-9851-0
29. Mead B, Tomarev S. Bone marrow-derived mesenchymal stem cells-derived exosomes promote survival of retinal ganglion cells through miRNA-dependent mechanisms. Stem Cells Transl Med. 2017;6(4):1273–1285. doi:10.1002/sctm.16-0428
30. Xu Y, Huang Y, Guo Y, et al. microRNA-690 regulates induced pluripotent stem cells (iPSCs) differentiation into insulin-producing cells by targeting Sox9. Stem Cell Res Ther. 2019;10(1):59. doi:10.1186/s13287-019-1154-8
31. Jeon K, Lim H, Kim J, et al. Differentiation and transplantation of functional pancreatic beta cells generated from induced pluripotent stem cells derived from a type 1 diabetes mouse model. Stem Cells Dev. 2012;21(14):2642–2655. doi:10.1089/scd.2011.0665
32. Kondo Y, Toyoda T, Ito R, et al. Identification of a small molecule that facilitates the differentiation of human iPSCs/ESCs and mouse embryonic pancreatic explants into pancreatic endocrine cells. Diabetologia. 2017;60(8):1454–1466. doi:10.1007/s00125-017-4302-7
33. Mandal P, De D, Yun K, et al. Improved differentiation of human adipose stem cells to insulin-producing β-like cells using PDFGR kinase inhibitor Tyrphostin9. Biochem Biophys Res Commun. 2020;533(1):132–138. doi:10.1016/j.bbrc.2020.08.090
34. Li D, Zhang W, Chen X, et al. Proteomic profiling of MIN6 cell-derived exosomes. J Proteomics. 2020;224:103841. doi:10.1016/j.jprot.2020.103841
35. Mandal P, De D, Im D, et al. Exosome-mediated differentiation of mouse embryonic fibroblasts and exocrine cells into β-like cells and the identification of key miRNAs for differentiation. Biomedicines. 2020;8(11):485. doi:10.3390/biomedicines8110485
36. Pan F, Brissova M. Pancreas development in humans. Curr Opin Endocrinol Diabetes Obes. 2014;21(2):77–82. doi:10.1097/MED.0000000000000047
37. Ray J, Kener K, Bitner B, et al. Nkx6.1-mediated insulin secretion and β-cell proliferation is dependent on upregulation of c-Fos. FEBS Lett. 2016;590(12):1791–1803. doi:10.1002/1873-3468.12208
38. Guay C, Menoud V, Rome S, et al. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun Signal. 2015;13:17. doi:10.1186/s12964-015-0097-7
39. Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi:10.1038/ncb1596
40. Gon Y, Maruoka S, Inoue T, et al. Selective release of miRNAs via extracellular vesicles is associated with house-dust mite allergen-induced airway inflammation. Clin Exp Allergy. 2017;47(12):1586–1598. doi:10.1111/cea.13016
41. Zhang J, Zhang F, Didelot X, et al. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genom. 2009;10:478. doi:10.1186/1471-2164-10-478
42. Garavelli S, Bruzzaniti S, Tagliabue E, et al. Blood co-circulating extracellular microRNAs and immune cell subsets associate with type 1 diabetes severity. Int J Mol Sci. 2020;21(2):477. doi:10.3390/ijms21020477
43. Xu Y, Zhang J, Fan L, et al. miR-423-5p suppresses high-glucose-induced podocyte injury by targeting Nox4. Biochem Biophys Res Commun. 2018;505(2):339–345. doi:10.1016/j.bbrc.2018.09.067
44. Yang W, Wang J, Chen Z, et al. NFE2 induces miR-423-5p to promote gluconeogenesis and hyperglycemia by repressing the hepatic FAM3A-ATP-Akt pathway. Diabetes. 2017;66(7):1819–1832. doi:10.2337/db16-1172
45. Hashemi J, Kajbafzadeh A, Ghorbani F, et al. Application of iPSCs derived pancreatic β-like cells using pancreatic bio-scaffold. Exp Cell Res. 2021;405(2):112667. doi:10.1016/j.yexcr.2021.112667
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.