Back to Journals » OncoTargets and Therapy » Volume 12
Exosome-Mediated Transfer of lncRNA HOTTIP Promotes Cisplatin Resistance in Gastric Cancer Cells by Regulating HMGA1/miR-218 Axis
Authors Wang J, Lv B, Su Y, Wang X, Bu J , Yao L
Received 20 September 2019
Accepted for publication 26 November 2019
Published 20 December 2019 Volume 2019:12 Pages 11325—11338
DOI https://doi.org/10.2147/OTT.S231846
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Jingyu Wang,1,* Baojun Lv,1,* Yonghui Su,1 Xiao Wang,1 Juyuan Bu,1 Lan Yao2
1Department of Gastrointestinal Surgery, The Fifth Affiliate Hospital of Sun Yat-Sen University, Zhuhai, People’s Republic of China; 2Department of Emergency Medicine, The Fifth Affiliate Hospital of Sun Yat-Sen University, Zhuhai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Juyuan Bu
Department of Gastrointestinal Surgery, The Fifth Affiliate Hospital of Sun Yat-Sen University, No. 52, Meihua East Road, Zhuhai, Guangdong 519000, People’s Republic of China
Email [email protected]
Lan Yao
Department of Emergency Medicine, The Fifth Affiliate Hospital of Sun Yat-Sen University, No. 52, Meihua East Road, Zhuhai, Guangdong 519000, People’s Republic of China
Email [email protected]
Background: Chemoresistance has become a major obstacle for cancer therapy in clinic. Long noncoding RNAs (lncRNAs) have been reported to play critical roles in the development of chemoresistance in various tumors, including gastric cancer (GC). However, the role of HOXA transcript at the distal tip (HOTTIP) within extracellular vesicles (exosomes) in cisplatin-resistant GC cells remains largely unknown.
Materials and methods: Cell proliferation, migration and invasion were detected using Cell Counting Kit-8 (CCK-8) and transwell assays, respectively. Western blot assay was employed to analyze the protein levels of E-cadherin, N-cadherin, Vimentin, CD63, CD83, GRP78, HMGA1, and high-mobility group A1 (HMGA1). The expression levels of HOTTIP, microRNA-218 (miR-218) and HMGA1were measured by quantitative real-time polymerase chain reaction (qRT-PCR). The interaction between miR-218 and HOTTIP or HMGA1 was predicted by bioinformatics software and confirmed by the dual-luciferase reporter and RNA immunoprecipitation (RIP) assays.
Results: Cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) were promoted in cisplatin-resistant GC cells. HOTTIP level was upregulated in cisplatin-resistant GC cells and its downregulation enhanced cisplatin sensitivity. Moreover, extracellular HOTTIP could be incorporated into exosomes and transmitted to sensitive cells, thus disseminating cisplatin resistance. Additionally, exosomal HOTTIP promoted cisplatin resistance via activating HMGA1 in GC cells. Interestingly, HMGA1 was a target of miR-218 and miR-218 could directly bind to HOTTIP. Clinically, high expression of exosomal HOTTIP in serum was associated with poor response to cisplatin treatment in GC patients.
Conclusion: Exosomal HOTTIP contributed to cisplatin resistance in GC cells by regulating miR-218/HMGA1 axis, providing a novel avenue for the treatment of GC.
Keywords: gastric cancer, cisplatin resistance, HOTTIP, exosomes, HMGA1, miR-218
Introduction
Gastric cancer is a frequent malignant tumor, ranking as the second leading cause of cancer-related death worldwide.1 Although advances in therapeutic during past decades, the prognosis for patients with GC is still poor, and 5-year survival rate is less than 25%.2,3 Cisplatin (DDP)-based chemotherapy is still a primary strategy for GC treatment; however, tumor recurrence commonly occurs after the development of cisplatin resistance.4,5 Hence, it is especially important to further study molecular mechanisms underlying of cisplatin resistance for improving GC treatment.
Exosomes are small membrane vesicles that originate from endosomal multivesicular bodies, with diameters ranging from 40 nm to 100 nm.6 In recent years, exosomes have drawn great attention in the field of biomarker discovery. Emerging evidence has demonstrated that cancer-derived exosomes promote cancer progression and metastasis in the tumor microenvironment.7 The emerging evidence has shown that exosomes from chemosensitive or resistant cells may potentially influence the therapeutic response via transferring specific genes, including long non-coding RNAs (lncRNAs).8,9 Thus, whether exosomes derived from cisplatin-resistant GC cells can confer cisplatin resistance to sensitive cells is worth further exploring.
LncRNAs, >200 nucleotides in length, lack protein-coding capacity.10 LncRNAs have been proven to serve as crucial regulators in the diverse biological processes through modulating genes expression at the post-transcriptional level.11–13 The lncRNA HOXA transcript at the distal tip (HOTTIP) is produced from the 5ʹ end of the HOXA cluster and its dysregulation was tightly associated with the development of human cancers.14 Moreover, HOTTIP is suggested to be expressed at a high level in various cancers, such as pancreatic cancer,15 lung cancer,16 hepatocellular carcinoma,17 colorectal cancer,18 and GC.19 Besides, HOTTIP has been identified to accelerate the development of cisplatin resistance in GC cells.20 Nevertheless, the exact biological role and underlying mechanism of exosomal HOTTIP from cisplatin-resistant GC cells need to be further investigated.
High-mobility group A1 (HMGA1), a new transcriptional regulator, is located at 6p21.31 and plays a key role in tumor progression.21 Recently, HMGA1 has been found to be expressed at a high level and acted as a tumor promoter in different cancers, including GC.22,23 It is well accepted that lncRNAs can serve as molecular sponges for miRNAs to modulate related gene expression. Bioinformatics analysis shows the potential binding sites between miR-218 and HOTTIP or HMGA1. Thus, we supposed that HOTTIP might influence cisplatin resistance through sponging miR-218 to regulate HMGA1expression.
In our study, the effect of exosome-transmitted HOTTIP on cisplatin resistance in GC cells was explored. Moreover, we probed the HOTTIP/miR-218/HMGA1 regulatory network and also explored the involvement of HOTTIP in the regulation of chemotherapeutic responses through tumor cell extracellular exosomes in GC.
Materials and Methods
Clinical Samples
A total of 58 serum samples were enrolled from the patients with GC who received cisplatin treatment at The Fifth Affiliate Hospital of Sun Yat-Sen University. The clinicopathological parameters of 58 patients are presented in Table 1. Briefly, venous blood (5 mL) from each patient was collected through venipuncture before chemotherapy began. Serum was separated by centrifugation (1600× g, 10 min, room temperature) within 2 h after collection. A new tube was used to transfer the supernatant, followed by centrifugation (12,000 × g, 10 min, 4°C) to discard the residual cells debris. Next, the RNase-free tubes were used to transfer the final supernatant, which was kept in −80°C freezer until RNA extraction. The patients with GC were divided into the response (complete response + partial response, 30 patients) group and non-response (stable disease + progressive disease, 28 patients) group in accordance with the Response Evaluation Criteria In Solid Tumors (RECIST) (version 1.1).24 Written informed consents have been acquired from all patients before blood collection. This research protocol was approved by Research Ethics Committee of The Fifth Affiliate Hospital of Sun Yat-Sen University.
 |
Table 1 The Clinicopathological Parameters of 58 Patients Who Accepted Cisplatin-Based Chemotherapy with Gastric Cancer |
Stability testing of exosomal HOTTIP in serum was carried out via treating the serum to various conditions, including incubation for different times (0 hr, 3 hrs, 6 hrs, 12 hrs and 24 hrs) at room temperature, RNase A digestion, and low (pH =1.0, HCl) or high (pH = 13.0, NaOH) pH solution for 3 h at room temperature, and measured by qRT-PCR to assess the HOTTIP level.
Cell Culture
Human GC cell lines (MGC-803 and MKN-45) were brought from Bena Culture Collection (Beijing, China). Both cell lines were maintained in RPMI 1640 (HyClone, Logan, UT, USA) with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA), penicillin (100 U/mL) and streptomycin (100 mg/mL) (Invitrogen, Carlsbad, CA, USA). Continuous exposure to stepwise-enhancing concentrations of cisplatin (Sigma-Aldrich, St. Louis, MO, USA) was used to establish cisplatin-resistant MGC-803 and MKN-45 cells. These cells were initially incubated in medium with a relatively low concentration of cisplatin (1 µM) in medium for 4 weeks. Subsequently, the surviving cells were exposed to a middle concentration of cisplatin (2 µM) for 6 weeks. Lastly, MGC-803 and MKN-45 cells were incubated in the culture medium containing a high concentration of cisplatin (5 µM). All cells were grown in a moist atmosphere with 5% carbon dioxide incubator at 37°C.
Cell Treatment and Transfection
RNase A and Triton X100 were bought from Qiagen (Waltham, MA, USA). RNase A (20 µg/mL, 1hr) was used to determine the change in the expression of HOTTIP. Triton X100 (0.1%, 30 min) was used to measure the existence of vesicles.
Small interfering RNA (siRNA) against HOTTIP (si-HOTTIP #1, si-HOTTIP #2 and si-HOTTIP #3), siRNA against HMGA1 (si-HMGA1), siRNA negative control (si-NC), pcDNA-HOTTIP overexpression plasmid (HOTTIP), empty pcDNA (pcDNA), miR-218 mimic (miR-218), and miR-negative control (miR-NC) were provided by RiboBio (Guangzhou, China). Transfection was performed with Lipofectamine 3000 (Invitrogen).
Exosomes Isolation
ExoQuick precipitation kit (System Biosciences, Mountain view, CA, USA) was used to extract exosomes from GC cells culture medium or serum samples. In brief, the culture medium and serum were unfrozen on ice, followed by centrifugation (3000 × g, 15 min) to discard the cell debris. Then, the supernatant (250 µL) was mixed with the ExoQuick exosome precipitation solution (63µL) and incubated at 4°Cfor 40 min. Next, the supernatant was carefully discarded by aspiration following centrifugation (1500 × g, 15 min), followed by another centrifugation (1500 × g, 5mins) to remove the residual liquid. Subsequently, phosphate buffered saline (PBS, 250 μL) was employed to resuspend the exosome containing pellet. Finally, the pellets containing exosomes were harvested for RNA isolations.
Subcellular Fractionation Location
Nuclear and cytosolic fractions were separated using the PARIS Kit (Life Technologies Corp., Grand Island, NY, USA). In brief, cells (MGC-803 and MKN-45) were washed using the PBS and placed on ice. After that, these cells were resuspended in fractionation buffer, followed by centrifugation (500 × g, 5 min, 4°C). Then, the cytoplasmic fraction was separated from the nuclear pellet and then cell fractionation buffer was used to wash the nuclear pellet. Subsequently, cell disruption buffer was employed to wash the nuclear pellet. Next, the sample was divided for RNA isolation. Finally, the expression of U6, GAPDH and HOTTIP was analyzed using qRT-PCR analysis. U6 was used as the nuclear control. And GAPDH was used as the cytoplasmic control.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Sangon Biotech, Shanghai, China) was applied to examine cell proliferation ability. Briley, growing cells at an exponential rate were sowed in 96-well plates (4000 cells/well). After treatment/transfection for 48 h, the culture medium was added with CCK-8 (10 μL) solution, then incubated for an additional 3 hrs at 37°C. Finally, the absorbance of test wells was read under a microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm.
Transwell Assay
Transwell chamber (8 μm pore size, Corning Inc., Corning, NY, USA) was applied to evaluate cell migration and invasion abilities. For migration assay, treated/transfected cells were resuspended in serum-free medium (RPMI 1640,100 μL) and placed in the upper chamber. Besides, RPMI 1640 with 10% FBS (500 μL) would be placed in the bottom chamber. Following incubation for 24 h, a cotton swab was applied to wipe the cells (located on the upper surface), and migrated cells (located on the bottom surface of the chamber) were fixed using 4% paraformaldehyde and stained using 0.5% crystal violet, and then counted by an inverted microscope (Olympus, Tokyo, Japan). For invasion assay, cells were placed into the upper chambers pre-coated with Matrigel (Becton Dickinson, San Jose, CA, USA). The remaining steps were employed in the like way as the migration assay.
Western Blot Assay
RIPA lysis buffer (Thermo Fisher Scientific) containing phenylmethanesulfonyl fluoride (PMSF; Beyotime, Shanghai, China) was utilized to extract the total protein. Bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology) was performed to determine the protein concentration. Then, the total protein in equal concentration was separated using 10–12% SDS-PAGE. After that, the gels containing related protein were transferred to a polyvinylidene fluoride (PVDF) membrane (0.2μm, Beyotime). Next, the membranes would be blocked using PBS containing 0.1% Tween-20 (PBST) and 5% non-fat dry milk, followed by incubation with specific primary antibody (4°C, overnight) against E-cadherin (1:500, ab15148, Abcam, Cambridge, UK), N-cadherin (1:500, ab18203, Abcam), Vimentin (1:1000, ab1373218, Abcam), CD63 (1:500, ab21630, Abcam), CD83 (1:1000, ab155760, Abcam), GRP78 (1:1000, ab32618 Abcam), GM130 (1:1000, ab32337, Abcam), HMGA1 (1:500, ab168260, Abcam), and GAPDH (1:2000, ab37168, Abcam). Following three washes with PBST, all membranes were incubated using secondary antibody (1:4000, Sangon Biotech) for 2 h at room temperature. Finally, enhanced chemiluminescence (ECL) system (Thermo Fisher Scientific) was performed to visualize the protein bands, and ImageJ software was used to evaluate the bands density. The protein levels were normalized by GAPDH.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA extraction from the exosome pellets was carried out by commercial miRNeasy Serum/Plasma kit (Qiagen). Total RNA from cells was extracted using Trizol reagent (Invitrogen). Reverse transcription reactions were conducted with the miScript II RTKit (Qiagen). Subsequently, qRT-PCR was carried out using SYBR green detection kit (Toyobo, Tokyo, Japan) on ABI 7500 real-time PCR system (Applied Biosystems). The sequences of primers were as follows: HOTTIP (forward, 5ʹ-AAGGAGGAAACGCCTTCTCG-3ʹ; Reverse, 5ʹ-CATCAAGCTAGGGTGGGGTG-3ʹ), miR-218 (Forward,5ʹ-GCGGCTTTGTGCTTGATCTAA-3ʹ; Reverse, 5ʹ-GTGCAGGGTCCGAGGT-3ʹ), HMGA1 (Forward, 5ʹ-AAGGGGCAGACCCAAAAA-3ʹ; Reverse, 5ʹ-TCCAGTCCCAGAAGGAAGC3ʹ), U6 (Forward,5ʹ-CTCGCTTCGGCAGCACA-3ʹ; Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ), GAPDH (forward, 5ʹ-GACTCCACTCACGGCAAATTCA-3ʹ; reverse, 5ʹ-TCGCTCCTGGAAGATGGTGAT-3ʹ). The levels of HOTTIP, miR-218, and HMGA1 were assessed using 2−ΔΔCt method. GAPDH and U6 levels were used for normalization.
Transmission Electron Microscopy (TEM)
The exosome pellets were resuspended in PBS (50 μL) and a drop of the suspension was placed on a sheet of parafilm. A carbon-coated copper grid was floated on the drop for 5 min at room temperature. After that, the grid was removed and excess liquid was drained via touching the grid edge against a piece of clean filter paper. The grid was then placed onto a drop of 2% phosphotungstic acid with pH 7.0 for approximately 5 s, and excess liquid was drained off. The grid was allowed to dry for several minutes and then examined through transmission electron microscopy.
Nanoparticle Tracking Analysis (NTA)
NTA was performed using a Nanosight LM10-HS (NanoSight, Amesbury, UK) to determine the concentration, size, and distribution of exosomes. Five recordings of 30 s each were captured, analyzed and the data from at least 5000 individual particle tracks were analyzed per sample. Data analysis was performed using NTA 2.1 software (NanoSight).
Dual-Luciferase Reporter Assay
Partial fragment of HOTTIP or 3ʹUTR of HMGA1 containing the predicted binding sites of miR-218 or mutant binding sites was amplified and cloned into the pmirGLO luciferase vector (Promega, Madison WI, USA), namely WT-HOTTIP, HMGA1 3ʹUTR-WT, MUT-HOTTIP, and HMGA1 3ʹUTR-MUT. The miR-NC or miR-218 was co-transfected with the reporter plasmid into MGC-803 and MKN-45 cells. Luciferase activity was evaluated with a dual-luciferase reporter assay system (Promega) following relevant transfection for 48 h.
RNA Immunoprecipitation (RIP) Assay
Magna RIP Kit (Millipore, Billerica, MA, USA) was used for RIP assay. In brief, cells (MGC-803 and MKN-45) were collected and resuspended in RNA immunoprecipitation lysis buffer with magnetic beads, and then incubated with anti-argonaute 2 (anti-Ago2) or IgG antibodies. After that, the protein was digested through proteinase K buffer, followed by RNA purification. Subsequently, the enrichment of HOTTIP, miR-218 and HMGA1 was determined by qRT-PCR.
Statistical Analysis
In this study, the data were displayed as the mean ± standard deviation (SD) with at least three independent experiments. The statistical differences between groups were assessed by Student’s t-test using GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA). Receiver operating characteristic (ROC) curves and the area under the curve (AUC) were performed using MedCalc version 11.4.2.0 software (MedCalc Software bvba, Ostend, Belgium) to distinguish between GC patients who responded to cisplatin and those who did not respond to cisplatin. The difference was considered to be significant when P< 0.05.
Results
Cisplatin-Resistant GC Cells Exhibited Migration, Invasion, and Epithelial-Mesenchymal Transition (EMT) and Upregulated Level of HOTTIP
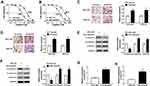
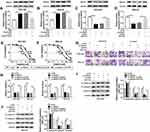
To explore the underlying regulatory mechanism of cisplatin resistance, we established cisplatin-resistant cell lines. To obtain cisplatin-resistant cells, treatment of MGC-803 and MKN-45 cells with gradually increasing doses of cisplatin in successive passages. Firstly, we determined the cell viability in cisplatin-resistant and cisplatin-sensitive cell lines (MGC-803 and MKN-45) exposed to different concentrations of cisplatin for 48 h using CCK-8 assay. As presented in Figure 1A and B, compared with the parental cells (MGC-803 and MKN-45), cell viability and IC50 value were increased in resistant cells (MGC-803/DDP and MKN-45/DDP), suggesting that resistant cells respond less to cisplatin treatment. Next, we used the transwell assay to assess cell migration and invasion capacities. As presented in Figure 1C and D, cell migration and invasion abilities were dramatically increased in MGC-803/DDP and MKN-45/DDP cells compared with that in MGC-803 and MKN-45 cells. Chemoresistant cells undergo EMT processes. Thus, EMT-related proteins were analyzed in cisplatin-resistant and cisplatin-sensitive cell lines by Western blot analysis. As displayed in Figure 1E and F, the protein levels of E-cadherin (an epithelial marker) were remarkably reduced and the expression levels of N-cadherin and Vimentin (mesenchymal markers) were increased in MGC-803/DDP and MKN-45/DDP cells relative to MGC-803 and MKN-45 cells. To test the effect of cisplatin resistance on the expression of HOTTIP, the HOTTIP expression was determined by qRT-PCR. As displayed in Figure 1G and H, the level of HOTTIP was markedly enhanced in resistant cells (MGC-803/DDP and MKN-45/DDP) in contrast to sensitive cells (MGC-803 and MKN-45). These results suggested that high expression of HOTTIP might be linked to cisplatin resistance in GC cells.
HOTTIP Downregulation Enhanced Cisplatin Sensitivity in Cisplatin-Resistant GC Cells
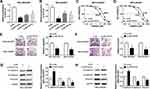
Based on the above findings, the effect of HOTTIP interference on cisplatin resistance in GC cells was further probed. Our results indicated that, compared within MGC-803/DDP and MKN-45/DDP cells transfected with si-NC, the expression level of HOTTIP was strikingly reduced in MGC-803/DDP and MKN-45/DDP cells transfected with si-HOTTIP#1, si-HOTTIP#2, and si-HOTTIP#3, especially in si-HOTTIP#1 group (Figure 2A and B). According to the knockdown efficiency, si-HOTTIP#1 was chosen for subsequent experiments. CCK-8 assay indicated that interference of HOTTIP resulted in significant inhibition of cell proliferation and decrease of IC50 values in MGC-803/DDP and MKN-45/DDP cells (Figure 2C and D). Additionally, cell migration and invasion capacities after silencing HOTTIP were analyzed using transwell assay. As shown in Figure 2E and F, the depletion of HOTTIP conspicuously inhibited migration and invasion abilities in MGC-803/DDP and MKN-45/DDP cells. Besides, inhibition of HOTTIP led to the obvious promotion of E-cadherin expression and reduction of N-cadherin and Vimentin abundances in MGC-803/DDP and MKN-45/DDP cells (Figure 2G and H). Taken together, our data demonstrated that HOTTIP downregulation sensitized MGC-803/DDP and MKN-45/DDP cells to cisplatin.
HOTTIP Was Secreted Through Incorporation into Exosomes in Cisplatin-Resistant GC Cells
Exosomes act as a crucial mediator of intercellular communication. To confirm whether HOTTIP was secreted by packaging into exosomes, the abundance change of extracellular HOTTIP was measured following treatment with RNase or RNase + Triton X100. As displayed in Figure 3A, HOTTIP expression in culture medium was little affected through the treatment of RNase alone but notably reduced when simultaneously treated with RNase and Triton X100, indicating that extracellular HOTTIP was protected by membrane rather than directly secreted. Exosomes from culture medium were purified and extracted to validate the results. The morphology and size of the exosomes were displayed in Figure 3B and C. Western blot assay showed that the exosomal protein markers (CD63 and CD81) were effectively expressed in exosomes rather than in cell extracts, and the protein levels of GAPDH, GRP78 (a major endoplasmic reticulum chaperone), GM130 (a cis-Golgi matrix protein) were mainly expressed in cell extracts rather than in exosomes (Figure 3D), suggesting that the extraction of exosomes was successful. Subsequently, we explored whether HOTTIP was incorporated into exosomes. ExoQuick purification kit (System Biosciences) was used to isolate exosomes from culture medium, and the exosomes were then subjected to qRT-PCR. As anticipated, HOTTIP could be detected in exosomes, and the abundance of HOTTIP was prominently increased in culture medium collected from cisplatin-resistant cells compared with that in culture medium from parental cells (Figure 3E), suggesting that extracellular HOTTIP was secreted by incorporation into exosomes in GC cells.
Exosome-Mediated Transfer of HOTTIP Induced Cisplatin Resistance in GC Cells
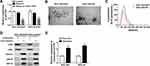
To further validate that exosomal HOTTIP was involved in cisplatin resistance, the MGC-803 and MKN-45 cells were incubated with PBS, exosomes (from MGC-803/DDP or MKN-45/DDP), exosomes + si-NC, or exosomes + si-HOTTIP#1. The qRT-PCR analysis showed that exosome treatment promoted HOTTIP expression in MGC-803 and MKN-45 cells (Figure 4A and B). However, knockdown of HOTTIP obviously reversed this effect. Thereby, we determined that HOTTIP-contained exosomes could be taken up by recipient cells. Next, we investigated whether MGC-803 and MKN-45 cells with high exosomal HOTTIP expression presented an increased resistance to cisplatin relative to control cells. Results revealed that both recipient cell lines showed a promoted cell proliferation ability following treatment of exosomes with respect to control cells, while these effects were abated by transfection with si-HOTTIP#1 (Figure 4C and D). Besides, cell migration and invasion were promoted by treatment of exosomes in MGC-803 and MKN-45 cells, whereas the promotive effect was abolished by inhibiting HOTTIP (Figure 4E and F). Moreover, the protein levels of N-cadherin and Vimentin were increased and E-cadherin level was reduced in MGC-803 and MKN-45 cells treated with exosomes in comparison with the cells treated with PBS. Similarly, these effects were reversed by knockdown of HOTTIP (Figure 4G and H). Collectively, these findings indicated that increased cisplatin-resistant potency was induced after treatment with exosomal HOTTIP.
Exosomal HOTTIP Promoted Cisplatin-Resistance by Activating HMGA1
HMGA1 has been confirmed to be involved in the progression of GC. After that, we probed whether the packaging of HOTTIP into exosomes was regulated by specific HMGA1. As anticipated, qRT-PCR assay showed that the protein abundance of HMGA1 was upregulated after treatment with exosomes in MGC-803 and MKN-45 cells, which was attenuated by knockdown of HOTTIP (Figure 5A and B). Besides, overexpression of HOTTIP abated the suppressive effect of HMGA1 knockdown on the expression of HMGA1 (Figure 5C and D). Moreover, upregulation of HOTTIP partly reversed anti-proliferation, anti-migration and anti-invasion effects caused by HMGA1 knockdown in GC-803 and MKN-45 cells (Figure 5E–H). In addition, HOTTIP overexpression abolished si-HMGA1-mediated promotion of E-cadherin level and reduction of N-cadherin and Vimentin expression in GC-803 and MKN-45 cells (Figure 5I and J). Thus, our results suggested that HMGA1 mediated the packaging of HOTTIP into exosomes.
Exosomal HOTTIP-Mediated HMGA1 Expression Through Functioning as a ceRNA for miR-218
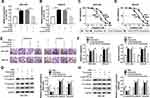
It is well known that lncRNAs may act as ceRNAs to modulate the target mRNAs of miRNAs. In order to verify this hypothesis, we firstly examined the localization of HOTTIP abundance in GC-803 and MKN-45 cells. The results proved that most of the HOTTIP were located in the cytoplasm, indicating that HOTTIP might regulate its targets at the post-transcriptional levels (Figure 6A and B). Subsequently, miRcode was employed to predict the potential target miRNAs for HOTTIP. Results proved that, miR-218, a well-studied tumor suppressor in several cancers, might be a possible target of HOTTIP (Figure 6C). Interestingly, starbase displayed that, HMGA1, a well-studied oncogene in some tumors, was a potential target of miR-218 (Figure 6C). Besides, results manifested that the abundance of miR-218 was obviously decreased in both cisplatin-resistant cells (MGC-803/DDP and MKN-45/DDP) compared with that in parental cells (MGC-803 and MKN-45) (Figure 6D). To certify the previous prediction, the data of dual-luciferase reports confirmed that co-transfection of WT-HOTTIP vector or HMGA13ʹUTR-WT with miR-218 mimic markedly decreased the activity of luciferase in MGC-803 and MKN-45 cells, whereas the luciferase activity was not changed in the cells co-transfected with MUT-HOTTIP or HMGA13ʹUTR-MUT vector and miR-218 mimic (Figure 6E–H). Moreover, RIP assay revealed that the enrichment of HOTTIP, miR-218 and HMGA1 immunoprecipitated with Ago2 was higher than the respective IgG group in MGC-803 and MKN-45 cells (Figure 6I and J). Besides, the protein expression of HMGA1 was enhanced in resistant cells (MGC-803/DDP and MKN-45/DDP) compared with the parental cells (MGC-803 and MKN-45) (Supplemental Figure 1A). Additionally, knockdown of HOTTIP increased the expression of miR-218 and its overexpression presented an opposite effect in MGC-803 and MKN-45 cells (Supplemental Figure 1B and 1C), suggesting that miR-218 was negatively by HOTTIP. Furthermore, Western blot proved that the protein level of HMGA1 was downregulated in MGC-803 and MKN-45 cells transfected with miR-218, which was reversed by co-transfection with HOTTIP (Figure 6K and L). These results demonstrated that HOTTIP acted as a sponge of miR-218 to regulate HMGA1 expression in GC cells.
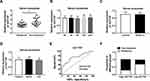
Serum Exosomal HOTTIP Was Associated with Cisplatin Resistance in GC Patients
Next, we measured the abundance of exosomal lncRNA HOTTIP in 58 serum samples from advanced patients receiving cisplatin treatment. According to Response Evaluation Criteria In Solid Tumors (RECIST) (version 1.1), the patients would be divided into the responding (30 patients) and non-responding (28 patients) groups. Results disclosed that the serum exosomal HOTTIP abundance was strongly upregulated in patients who did not respond to treatment with cisplatin in contrast to those who responded to cisplatin (Figure 7A). Since a better stability was a crucial prerequisite for tumor markers, HOTTIP stability in serum exosomes was then determined through exposing exosomes to various conditions, including incubation for different times (0 hr, 3 hrs, 6 hrs, 12 hrs and 24 hrs) at room temperature, RNase A digestion, and low (pH = 1.0) or high (pH = 13.0) pH solution at room temperature for 3 h. As shown in Figure 7B–D, the exosomal HOTTIP level was not notably affected by any of the experimental conditions, suggesting that exosomal HOTTIP was stable in serum exosomes. Moreover, we explored the diagnostic potential of HOTTIP by performing ROC analysis. The results showed that, the AUC, diagnostic sensitivity and specificity reached 0.743 (95% confidence interval, 0.6149–0.8708) with the established cut-offs (1.905) (Figure 7E). According to these stratification criteria (1.905), patients were divided into the low and high HOTTIP level groups, and the proportion of patients who did not respond to chemotherapy was obviously elevated in the high exosomal HOTTIP level group relative to the low abundance group (Figure 7F). Therefore, our data suggested that exosomal HOTTIP in serum might act as a promising diagnostic biomarker for the patients with GC.
Discussion
In spite of cisplatin-based chemotherapy is particularly effective in a large number of cancer cases, the development of chemotherapy resistance has become a main obstacle for the treatment of cancer patients.4,25 Increasing reports have revealed the molecular and cellular mechanisms of drug resistance in diverse tumors.26,27 Besides, exosomal lncRNAs have been considered to play vital roles in the chemoresistance of various cancers.28,29 In our research, the effects and mechanism of exosomal HOTTIP in cisplatin resistance were explored in GC cell. Our data suggested that HOTTIP abundance was elevated in cisplatin-resistant GC cells. Moreover, we found that exosomal HOTTIP might contribute to cisplatin resistance in GC cells by regulating HMGA1 expression via sponging miR-218.
It has been reported that abnormal expression of HOTTIP was tightly associated with the development of chemoresistance in different tumors. For instance, HOTTIP expression was increased in lung adenocarcinoma and acted as an oncogene to promote the drug resistance of lung adenocarcinoma cells.30 Li et al proved that HOTTIP level was also enhanced in osteosarcoma and its overexpression was tightly associated with chemoresistance in osteosarcoma.31 Here, we found that abundance of HOTTIP was increased in resistant GC cells, and its knockdown elevated cisplatin sensitivity in cisplatin-resistant GC cells, suggesting that HOTTIP was required for the retention of a cisplatin-resistant status in GC cells.
Exosomes have been identified to be involved in cellular communication and modulation of the chemosensitivity of the recipient cells.32 More and more reports have proved that RNAs (mRNAs, miRNAs and lncRNAs) are secreted from cancer cells into body fluids by exosomes.33,34 Besides, growing evidence has emphasized exosomes can protect lncRNAs from degradation in the circulation, and cancers at an early stage may be determined by exosomal lncRNAs.35,36 Previous study has shown that expression level of exosomal HOTTIP was typically upregulated in GC than in normal control and it might be a promising biomarker for GC in diagnosis and prognosis.37 In our research, we verified that lncRNA HOTTIP was incorporated into exosomes and could regulate cisplatin resistance. The expression of HOTTIP was rarely affected by exposure to RNase A alone in the culture medium, while its expression was greatly inhibited after treatment with RNase A and Triton X100, indicating that HOTTIP was contained in vesicles, including exosomes. In addition, we found that exosomal protein markers (CD63 and CD81) were effectively expressed in exosomes, and HOTTIP expression from the extracted exosomes was enhanced in cisplatin-resistant GC cells. Furthermore, we also demonstrated that the parental cells treated with exosomes containing HOTTIP could induce an elevated cisplatin resistance. Therefore, these findings indicated that exosomal HOTTIP promoted the development of cisplatin resistance in GC cells.
HMGA1 dysregulation has been confirmed to be associated with a drug-resistant phenotype.38,39 To explore whether the packaging of HOTTIP into exosomes was mediated by HMGA1, MGC-803 and MKN-45 cells were exposed to exosomes or/and si-HOTTIP. And we found that HMGA1 expression was upregulated after treatment with exosomes, while the effect was reversed by knockdown of HOTTIP. Besides, overexpression of HOTTIP abated the enhanced effect of HMGA1 knockdown on cisplatin sensitivity in GC cells. Numerous reports have demonstrated that lncRNAs might function as ceRNAs to modulate the target mRNAs of miRNAs.40,41 We wondered whether HOTTIP acted as ceRNAs to regulate cisplatin resistance in GC cells. Firstly, HOTTIP expression was localized in GC cells. Results revealed that HOTTIP was located in the cytoplasm, suggesting that HOTTIP might modulate its targets at the post-transcriptional levels. Interestingly, bioinformatics analysis predicted that putative binding sites between miR-218 and HOTTIP or HMGA1, and the prediction was confirmed through dual-luciferase reporter assay and RIP assay. MiR-218 has been confirmed to serve as a tumor suppressor in GC.42 Moreover, Western blot disclosed that the protein abundance of HMGA1 was decreased in GC cells transfected with miR-218, which was reversed by co-transfection with HOTTIP. These data suggested that HOTTIP regulated HMGA1 expression by sponging miR-218.
According to the functional observations, the exosomal HOTTIP abundance in serum samples of GC patients was measured by qRT-RCR. In recent years, lncRNAs in serum or plasma were often used as predictors in GC.43,44 However, these potential biomarkers are commonly observed at relatively fewer levels and are easily degraded through the complexity of bodily fluids. Previous studies stated that lncRNAs were enriched and stable in circulatory exosome system and could avoid being degraded by RNase.45,46 Most importantly, exosome proteins may reflect their cellular origin, which may help us detect cancers at an early stage.47 As expected, our results proved that the exosomal HOTTIP abundance was enhanced in cisplatin-resistant patients as well as stably expressed in various conditions, and was involved in cisplatin response. Thus, these data indicated that exosomal HOTTIP might be used for clinical diagnosis or prognosis prediction of GC.
In conclusion, these findings suggested that exosome-mediated transfer of HOTTIP induced cisplatin resistance by regulating miR-218/HMGA1 axis in GC cells, providing a promising exosome lncRNA-directed therapy for the patient with GC and a vital theoretical basis for GC therapy. Besides, HOTTIP/miR-218/HMGA1 axis might provide a possible new strategy for the treatment of other cancers in the future. However, the biological function of exosome HOTTIP in vivo remains unclear, which will be the focus of our future research.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fitzmaurice C, Dicker D, Pain A, Collaboration GBODC. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505. doi:10.1001/jamaoncol.2015.0735
2. Russo A, Li P, Strong VE. Differences in the multimodal treatment of gastric cancer: east versus west. J Surg Oncol. 2017;115(5):603–614. doi:10.1002/jso.24517
3. Durães C, Almeida GM, Seruca R, et al. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464(3):367–378. doi:10.1007/s00428-013-1533-y
4. Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869. doi:10.1038/onc.2011.384
5. Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27–36. doi:10.1016/j.phrs.2016.01.001
6. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi:10.1016/j.tcb.2015.01.004
7. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91(4):431–437. doi:10.1007/s00109-013-1020-6
8. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi:10.1016/j.ccell.2016.03.004
9. Xu C, Yang M, Ren Y, et al. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:4362–4368.
10. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
11. Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
12. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7. doi:10.1038/nrg3606
13. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861. doi:10.1038/nrg3074
14. Zhang H, Zhao L, Wang Y-X, et al. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumor Biol. 2015;36(11):8805–8809. doi:10.1007/s13277-015-3645-2
15. Li Z, Zhao X, Zhou Y, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13(1):84. doi:10.1186/s12967-015-0442-z
16. Deng H, Chen L, Fan T, et al. Long non-coding RNA HOTTIP promotes tumor growth and inhibits cell apoptosis in lung cancer. Cell Mol Biol (Noisy-Le-Grand). 2015;61(4):34–40.
17. Tsang FH, Au SL, Wei L, et al. Long non‐coding RNA HOTTIP is frequently up‐regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR‐125b. Liver Int. 2015;35(5):1597–1606. doi:10.1111/liv.12746
18. Ren Y-K, Xiao Y, Wan X-B, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11458.
19. Ye H, Liu K, Qian K. Overexpression of long noncoding RNA HOTTIP promotes tumor invasion and predicts poor prognosis in gastric cancer. Onco Targets Ther. 2016;9:2081.
20. Zhang X-W, Bu P, Liu L, et al. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462(3):227–232. doi:10.1016/j.bbrc.2015.04.121
21. Sumter T, Xian L, Huso T, et al. The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr Mol Med. 2016;16(4):353–393. doi:10.2174/1566524016666160316152147
22. Zhang X, Tao T, Liu C, et al. Downregulation of miR-195 promotes prostate cancer progression by targeting HMGA1. Oncol Rep. 2016;36(1):376–382. doi:10.3892/or.2016.4797
23. Akaboshi S-I, Watanabe S, Hino Y, et al. HMGA1 is induced by Wnt/β-catenin pathway and maintains cell proliferation in gastric cancer. Am J Pathol. 2009;175(4):1675–1685. doi:10.2353/ajpath.2009.090069
24. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi:10.1016/j.ejca.2008.10.026
25. Chen J, Solomides C, Parekh H, et al. Cisplatin resistance in human cervical, ovarian and lung cancer cells. Cancer Chemother Pharmacol. 2015;75(6):1217–1227. doi:10.1007/s00280-015-2739-2
26. Hu T, Li Z, Gao CY, et al. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22(30):6876–6889. doi:10.3748/wjg.v22.i30.6876
27. Adamska A, Elaskalani O, Emmanouilidi A, et al. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv Biol Regul. 2018;68:77–87. doi:10.1016/j.jbior.2017.11.007
28. Dong H, Wang W, Chen R, et al. Exosome-mediated transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol. 2018;53(3):1013–1026. doi:10.3892/ijo.2018.4467
29. Zhang W, Cai X, Yu J, et al. Exosome-mediated transfer of lncRNA RP11‑838N2. 4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53(2):527–538. doi:10.3892/ijo.2018.4412
30. Zhang G, Song W, Song Y. Overexpression of HOTTIP promotes proliferation and drug resistance of lung adenocarcinoma by regulating AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(24):5683–5690. doi:10.26355/eurrev_201712_14013
31. Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J Transl Res. 2016;8(5):2385.
32. Takahashi K, Yan IK, Kogure T, et al. Extracellular vesicle‐mediated transfer of long non‐coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4(1):458–467. doi:10.1016/j.fob.2014.04.007
33. Melo SA, Sugimoto H, O’connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi:10.1016/j.ccell.2014.09.005
34. Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi:10.1016/j.bbagen.2012.03.017
35. Yang H, Fu H, Xu W, et al. Exosomal non-coding RNAs: a promising cancer biomarker. Clin Chem Lab Med. 2016;54(12):1871–1879. doi:10.1515/cclm-2016-0029
36. Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015;36(3):2007–2012. doi:10.1007/s13277-014-2807-y
37. Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17(1):68. doi:10.1186/s12943-018-0817-x
38. Kim DK, Seo EJ, Choi EJ, et al. Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Exp Mol Med. 2016;48(8):e255. doi:10.1038/emm.2016.73
39. Liau S, Jazag A, Ito K, et al. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007;96(6):993. doi:10.1038/sj.bjc.6603654
40. Jalali S, Bhartiya D, Lalwani MK, et al. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi:10.1371/journal.pone.0053823
41. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi:10.1016/j.bbagrm.2015.06.015
42. Tie J, Pan Y, Zhao L, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6(3):e1000879. doi:10.1371/journal.pgen.1000879
43. Zhou X, Yin C, Dang Y, et al. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi:10.1038/srep11516
44. Fu M, Huang Z, Zang X, et al. Long noncoding RNA LINC 00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2018;51(1):e12425. doi:10.1111/cpr.2018.51.issue-1
45. Gezer U, Zgür E, Cetinkaya M, et al. Long non‐coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi:10.1002/cbin.10301
46. Zhou R, Chen KK, Zhang J, et al. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer. 2018;17(1):75. doi:10.1186/s12943-018-0823-z
47. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi:10.1172/JCI81135
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.