Back to Journals » International Journal of Nanomedicine » Volume 18
Exosome-Delivered circSTAU2 Inhibits the Progression of Gastric Cancer by Targeting the miR-589/CAPZA1 Axis
Authors Zhang C, Wei G, Zhu X, Chen X, Ma X, Hu P, Liu W, Yang W, Ruan T, Zhang W, Wu C, Tao K
Received 7 October 2022
Accepted for publication 22 December 2022
Published 6 January 2023 Volume 2023:18 Pages 127—142
DOI https://doi.org/10.2147/IJN.S391872
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Chenggang Zhang,* Guanxin Wei,* Xiuxian Zhu,* Xiang Chen, Xianxiong Ma, Peng Hu, Weizhen Liu, Wenchang Yang, Tuo Ruan, Weikang Zhang, Chuanqing Wu, Kaixiong Tao
Department of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Kaixiong Tao; Chuanqing Wu, Department of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, No. 1277 Jiefang Avenue, Wuhan, Hubei Province, 430022, People’s Republic of China, Tel +86 13507155452 ; +86 13995598966, Email [email protected]; [email protected]
Background: Circular RNAs (circRNAs) are endogenous noncoding RNAs that play vital roles in many biological processes, particularly in human cancer. Recent studies indicate that circRNAs play an important role in tumor progression through exosomes. However, the specific functions of gastric cancer-derived exosomes and the role of circSTAU2 in gastric cancer (GC) remain largely unknown.
Methods: Differentially expressed circRNAs in GC were identified by circRNA microarrays analysis and quantitative real-time polymerase chain reaction (qRT-PCR). The role of circSTAU2 in GC was verified by circSTAU2 knockdown and overexpression with functional assays both in vitro and in vivo. Fluorescence in situ hybridization (FISH), immunofluorescence, RNA immunoprecipitation (RIP), dual-luciferase reporter assay, qRT-PCR and Western blot were adopted to evaluate the expression and regulatory mechanism of MBNL1, circSTAU2, miR-589 and CAPZA1. Furthermore, the role of exosomes was demonstrated by transmission electron microscopy and nano-sight particle tracking analysis.
Results: CircSTAU2, mainly localized in the cytoplasm, was significantly downregulated in GC. CircSTAU2 overexpression inhibited GC cell proliferation, invasion and migration both in vitro and in vivo, while circSTAU2 knockdown had the inverse effect. CircSTAU2 could be wrapped in exosomes and delivered to recipient cells, and functioned as a sponge for miR-589 to relieve its inhibitory effect on CAPZA1, thus inhibiting GC progression. Furthermore, MBNL1 acted as the upstream RNA-binding protein of circSTAU2 and significantly influenced the circularization and expression of circSTAU2.
Conclusion: Exosome-delivered circSTAU2 may act as a tumor suppressor that restrains GC progression via miR-589/CAPZA1 axis, which demonstrates a potential therapeutic target for GC.
Keywords: exosomes, circ-RNAs, bioinformatics, gastric cancer, cancer progression
Introduction
Gastric cancer (GC) has the fifth most prevalent occurrence rate in the world.1 Since early clinical symptoms of GC are not obvious, it is often at an advanced stage when diagnosed, which makes it the fourth leading cause of global cancer death.1 GC is a highly heterogeneous disease both molecularly and phenotypically. Complex interactions between Helicobacter pylori infection and environmental, microbial, and host genetic factors can lead to GC.2–4 Although advanced GC is treated by surgical resection and chemotherapy, the survival rate remains poor.5 Therefore, it is of great significance to explore the specific mechanism that contributes to GC progression.
Circular RNAs (circRNAs) are formed by precursor mRNA exon back-splicing in eukaryotes.6,7 In circRNAs, the downstream splice-donor site can covalently attach to an upstream splice-acceptor site; hence, it is shaped like a single-stranded ring.8 More and more studies show that circRNAs are involved in many critical biological functions, particularly in the progression of human cancers.9,10 To illustrate, Wang et al showed that circACTN4 facilitates the pathogenesis and progression of breast cancer.11 Wang et al demonstrated that circSPARC promotes the migration and proliferation of colorectal cancer.12 However, the specific role of circSTAU2 in cancer progression, especially in GC, remains poorly understood.
CircRNAs are involved in tumor progression via multiple mechanisms, the best-known of which is through direct binding to miRNAs as miRNA sponge.13 For instance, Chen et al showed that circDLG1 enhances CXCL12 expression by sponging miR-141-3p, which promotes proliferation and migration of GC.14 Recent studies have shown that circRNAs can also perform functions by promoting the transcription of parental genes15 or interacting with proteins.16
Exosomes are nanoscale membrane vesicles with diameters ranging from 30 to 200 nm that play vital roles in intercellular communications, which participate in many physiological and pathological processes.17 Tumor-derived exosomes play an important role in tumor progression.18 In addition, an increasing number of studies have shown that circRNAs are enriched in exosomes and regulate tumor progression through the transmission of exosomes between cells.19–21 However, how exosomes-delivered circRNAs are involved in the regulation of GC progression remains elusive.
In our study, we found that hsa_circ_0001811 (referred to as circSTAU2) has a lower expression in GC and is important for the suppression of GC progression. Mechanistically, circSTAU2 increases CAPZA1 expression by acting as a sponge for miR-589. Moreover, we found that circSTAU2 can be wrapped in exosomes and transmit the inhibitory effect on GC progression, which demonstrates a potential idea for GC therapy.
Materials and Methods
Bioinformatics Analysis
A thorough search for available gastric circRNA datasets was performed with the Gene Expression Omnibus (GEO) database (accession numbers: GSE93541 and GSE184882). DECs between GC and normal tissues were identified with the “MetaDE” package.22 GC miRNAs data were acquired from The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) miRNA database. The “limma” package23 was adopted to screen differentially expressed miRNAs (DEMs) between GC and normal tissues. The DEMs were subsequently overlapped with the predicted miRNAs from the CircInteractome database. GEO datasets (accession numbers: GSE64951 and GSE63089) were adopted to search for the differentially expressed genes (DEGs) between GC and normal tissues. The DEGs were then overlapped with the predicted target genes of miRNA from the starBase and TargetScan databases. RBPdb and RBPmap databases were employed to search for potential RNA-binding proteins (RBPs) of our target circRNA.
Patients and Tissue Samples
Fifty-six tumors and adjacent normal samples were acquired from GC patients who underwent surgery at the Department of Gastrointestinal Surgery, Union Hospital of Huazhong University of Science and Technology (HUST). All procedures were approved by the Ethics Committee of Union Hospital, HUST and performed according to the principles of the Declaration of Helsinki.
Cell Culture
The GC cell lines MKN45 and AGS were bought from Procell (Wuhan, China). SGC7901 and BGC823 were bought from Beyotime (Shanghai, China). MGC803 and the gastric mucosal epithelial cell line GES-1 were purchased from iCell Bioscience (Shanghai, China), all of which have been authenticated by DNA short tandem repeat test. RPMI-1640 medium with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) was used to culture the cells.
Gene Overexpression and Knockdown
Oligonucleotides of miR-589 mimics, CAPZA1-short hairpin RNA (shRNA), MBNL1-siRNA, KHDRBS3-siRNA, and negative control (NC) were synthesized by GeneChem (Shanghai, China). The MBNL1 plasmid (GeneChem, Shanghai, China) was used for MBNL1 overexpression. A specific shRNA for circSTAU2 was inserted into GV298 (GeneChem, Shanghai, China) to knock down the expression of circSTAU2. Oligonucleotides encoding the circSTAU2 sequence were synthesized by TSINGKE (Wuhan, China) and cloned into pLCDHciR (GeneChem, Shanghai, China) for circSTAU2 overexpression. The sequences are available in Supplementary Table S1. Lipofectamine 3000 (Invitrogen, MA, USA) was employed to transfect plasmids or siRNAs.
Quantitative Real‑time Polymerase Chain Reaction (qRT‑PCR)
TRIzol reagent (Invitrogen) was used to extract total RNA from cells or tissues as the manufacturer’s instructions. The total RNA was then reversely transcribed into cDNA using PrimeScript RT Master Mix (TaKaRa, Japan). The SYBR Green PCR kit (TaKaRa, Kyoto, Japan) was employed to conduct qRT-PCR with ABI StepOne Plus system (Thermo Fisher Scientific, USA). GAPDH and U6 were adopted as internal controls. 2−ΔΔCT method was used to calculate the relative expression. The sequences of primers are listed in Supplementary Table S2.
Western Blot (WB) Analysis
WB analyses were performed as previously described.24 Briefly, RIPA lysis buffer was used to extract the total proteins of cells, which were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto PVDF membranes. 5% milk in TBST buffer was used to block the membranes for 1 h before they were incubated with primary antibodies overnight. Enhanced chemiluminescence system (Millipore) was employed to detect the signal of the membranes after they were incubated with second antibody and washed with TBST for 3 times. Detailed antibody information is available in Supplementary Table S3.
Colony Formation Assays
Six-well plates were planted with 1000 cells per well and cultured for 2 weeks in complete medium. 0.1% crystal violet was used for staining of the cell colonies.
Cell Counting Kit 8 (CCK8) Assay
96-well plates were planted with 2×103 cells per well. At scheduled time points, the cells were incubated with CCK-8 reagent (Dojindo TLaboratories, Kumamoto, Japan) for 2 h, and the OD value was detected at 450 nm.
Transwell Assay
The transwell assays were performed as previously described.25 The cells with 200 μL serum-free medium were planted on the upper chamber, and 600 μL medium with 25% FBS was added to the lower chamber. After incubation for 24 h, the cells invading to the bottom side of the membrane were fixed with paraformaldehyde and stained with 0.1% crystal violet.
Flow Cytometry
Cell apoptosis was evaluated by flow cytometry. After transfected for 48 h, the GC cells were harvested and subjected to apoptosis assays, which were conducted with apoptosis assay kit (BD Pharmingen, NJ, USA) following the manufacturer’s protocol. The samples were finally analyzed by Flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
RNA Immunoprecipitation (RIP)
RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) was adopted for RIP assays according to the manufacturer’s instructions. Cell lysates were incubated with protein A/G agarose beads and MBNL1 antibody (1:50, #94633, CST) or IgG overnight at 4 ℃. Then the precipitates were washed and co-precipitated RNA was isolated, which was detected by qRT-PCR. And the reversely transcribed products of co-precipitated RNA were verified by DNA electrophoresis.
RNA-Fluorescence in situ Hybridization (FISH) Assay
The RNA-FISH kit (C10910, RiboBio, China) was employed to conduct RNA-FISH assay as the instructions of manufacturer. Probes targeting circSTAU2 (sense: 5’-CATGTACAATCAGAGATGTTCTCAGTGCAG-3’; antisense: 5’-CTGCACTGAGAACATCTCTGATTGTACATG-3’) labeled with biotin were synthesized by RiboBio Technology Co., Ltd. Zeiss confocal laser scanning microscope (LSM 780 with Airyscan; Carl Zeiss, Germany) was employed to measure the fluorescence.
Immunofluorescence
Cell immunofluorescence experiments were performed to detect the localization and expression level of MBNL1 as previously described.24 GC cells were washed with Phosphate-buffered saline (PBS) for 3 times after 48 h of transfection, which were fixed with −20℃ ethanol for 10 min and permeabilized with 0.5% triton-X100 for 10 min. Then the cells were blocked with 10% goat serum for 0.5 h before incubated with MBNL1 (1:50, 66837-1-Ig, Proteintech) at 4℃ overnight. The second day, the cells were incubated with secondary antibody for 1 h and DAPI for 5 min. The images were acquired with Zeiss confocal laser scanning microscope (LSM 780 with Airyscan; Carl Zeiss, Germany).
Dual-Luciferase Reporter (DLR) Assay
GC cells were co-transfected with WT or Mut circSTAU2, CAPZA1 3’UTR, and miR-589 mimics or NC mimics. After 48 h of transfection, the cells were subjected to dual-luciferase assay, and the DLR kit (TransGene, Beijing, China) was used to measure the luciferase activity. Renilla luciferase (RLUC) activity was used as internal control for Firefly luciferase (FLUC). Relative luciferase activity was calculated by the ratio of FLUC to RLUC.
Exosomes Isolation and Analysis
Medium with 10% exosome-removed FBS (Vivacell, Shanghai, China) was adopted to culture the cells. The supernatants of cells were collected to extract exosomes after cultured for 48 h. Firstly, the supernatants were differentially centrifuged at 500 × g and 3000 × g for 10 min, which were then subjected to the filtration with 0.2 μm filters and centrifuged at 120,000 × g at 4 ℃ for 2 h. PBS was used to resuspend and collect the exosomes. Transmission electron microscopy (Thermo, Waltham, MA, USA) was employed to photograph the exosomes. NanoSight NS300 system (NanoSight Technology, Malvern, UK) was employed to track the sizes and numbers of exosomes. Exosome samples were exposed to a laser, with their tracks analyzed by nanoparticle tracking analysis software (version 2.3).
Exosomes Labeling and Photographing
PKH26 membrane dye (PKH26GL, Sigma, Germany) was used to stain the exosomes. 12 mg of labeled exosomes were co-cultured with 1×105 cells for 3 h, which were then fixed and dyed with DAPI and photographed with a Zeiss confocal laser scanning microscope (LSM 780 with Airyscan; Carl Zeiss).
In vivo Xenograft Assay
An in vivo xenograft assay was conducted using male BALB/c nude mice (4 weeks old) purchased from Huafukang (Beijing, China), which were randomly separated into two groups. 100 μL PBS with 5×106 cells were injected into the groin of mice to develop subcutaneous xenograft models. Tumor volumes were calculated by tumor length and width, which were recorded every 4 d after implantation. After 4 weeks, all mice were sacrificed and the xenografts were extracted.
Statistical Analysis
All in vitro assays were performed in triplicate. Data are presented as the mean ± standard deviation (SD), and statistical analyses were conducted with GraphPad Prism 8 or SPSS 26.0 software. Student’s t-test was adopted to compare the difference. P < 0.05 was considered statistically significant.
Results
CircSTAU2 is Downregulated in GC
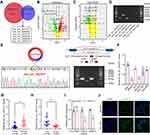
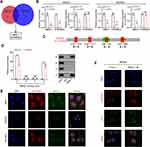
The microarray datasets GSE93541 and GSE184882 were used to explore DECs between GC and normal tissues. After combining the data of the two datasets, six DECs were selected for following analyses (Figure 1A–C). The PCR products of these six DECs were validated by DNA electrophoresis. The results showed three clearly visible strips, which suggested the possible existence of these three circRNAs in gastric cancer cells (Figure 1D). We further conducted sanger sequencing on the PCR products of these three circRNAs and found that only hsa_circ_0001811 was consistent with the sequence information in circbase database, and the junction of exon 4 and exon 5 formed a cyclization site. DNA electrophoresis verified the circular structure of hsa_circ_0001811 (Figure 1E). We then detected the expression of hsa_circ_0001811 in GC cell lines and specimens using qRT-PCR. The results revealed that hsa_circ_0001811 was consistently downregulated in GC cell lines and tumor tissues compared with GES-1 and normal tissues (Figure 1F and G). Moreover, by analyzing the correlation between hsa_circ_0001811 expression and clinicopathological information, we found that TNM III–IV stage gastric cancer patients had a significantly lower expression of hsa_circ_0001811 (Figure 1H). As shown in Figure 1F, hsa_circ_0001811 was most significantly downregulated in MKN45 and BGC823 cells. Therefore, we selected these two cell lines for further study and named hsa_circ_0001811 as circSTAU2. Resistance to RNase R exonuclease digestion suggested that circSTAU2 had a circular RNA structure (Figure 1I). FISH assay showed that endogenous circSTAU2 was mainly enriched in the cytoplasm (Figure 1J).
CircSTAU2 Suppresses GC Cell Proliferation, Invasion and Migration in vitro and in vivo
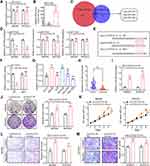
To investigate the specific role of circSTAU2 in human GC, we transfected MKN45 and BGC823 cells with overexpression plasmid or shRNA. The results indicated that the expression of circSTAU2 in MKN45 and BGC823 cells was altered by successful transfection (Figure 2A). CCK8 and colony formation assays revealed that circSTAU2 overexpression significantly inhibited the proliferation of GC cells (Figure 2B and C). Transwell assays showed that the invasive and migratory capacities of MKN45 and BGC823 cells were suppressed by circSTAU2 overexpression (Figure 2D and E). On the contrary, circSTAU2 knockdown had the opposite effects on GC cells. In addition, we found that enhancing the expression of circSTAU2 could promote apoptosis, whereas the opposite result was obtained by reducing circSTAU2 expression (Figure 2F). Furthermore, MKN45 cells with a stable overexpression of circSTAU2 were subcutaneously injected into nude mice, resulting in an obvious decline in the growth of xenograft tumors (Figure 2G). These results confirmed that circSTAU2 can suppress GC cell proliferation, invasion and migration.
CircSTAU2 Functions as a Sponge for miR-589 in GC
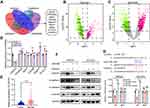
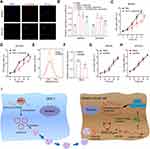
Previous reference indicates that circRNAs can function as regulators of the transcription of its parental genes or serve as a competitive endogenous RNA to sponge miRNAs.26 To determine the potential mechanism of circSTAU2, we overexpressed circSTAU2 and examined whether it affected the expression of STAU2. qRT-PCR results showed that the STAU2 expression had no significant change (Figure 3A). However, the RIP assay with AGO2 indicated that circSTAU2 was significantly enriched, suggesting that circSTAU2 may serve as a miRNA sponge (Figure 3B). We then used the TCGA and CircInteractome databases to predict the target miRNAs of circSTAU2. By combining the results, three miRNAs were selected (Figure 3C). qRT-PCR showed that only miR-589 expression in GC cells was affected by circSTAU2 overexpression (Figure 3D). By the starbase database, we found a binding site between circSTAU2 and miR-589 (Figure 3E). The DLR assay revealed that compared with circSTAU2 with a mutant binding site, the miR-589 mimics reduced the relative luciferase intensity of circSTAU2 with a wild-type binding site, which confirmed the interaction between circSTAU2 and miR-589 through the binding site (Figure 3F). The expression level of miR-589 was detected and was found significantly increased in GC cells and tissues (Figure 3G and H). To understand whether circSTAU2 suppressed GC progression by binding with miR-589, we conducted functional experiments with co-transfection of miR-589 mimics and circSTAU2 overexpression plasmid into GC cells. We found that miR-589 mimics could rescue the proliferative, invasive and migratory capacities of GC cells, which were suppressed by circSTAU2 overexpression (Figure 3I–M). These results indicated that circSTAU2 inhibits GC progression by acting as a sponge for miR-589.
CAPZA1 is Directly Targeted by miR-589
To identify the target genes of miR-589, we analyzed the DEGs between GC and normal tissues from the GSE64951 and GSE63089 datasets, which were overlapped with the predicted target genes of miR-589 from the starBase and TargetScan databases (Figure 4A–C). Six potential target genes were identified. qRT-PCR results showed that only CAPZA1 was consistently downregulated in GC cells (Figure 4D), and the expression of CAPZA1 in GC tissues was lower than that in normal tissues (Figure 4E). Furthermore, previous study showed that CAPZA1 was negatively associated with epithelial–mesenchymal transition (EMT)-associated markers.27 Our results suggested that the enhanced EMT process by miR-589 mimics could be reversed by CAPZA1 overexpression (Figure 4F). The DLR assay confirmed that miR-589 could bind to the 3′UTR of CAPZA1 (Figure 4G). These results indicated that miR-589 directly targets CAPZA1.
CircSTAU2 Overexpression Rescues the Effects of CAPZA1 Knockdown
Next, we performed rescue assays to explore the relationship between CAPZA1 and circSTAU2 as well as their effects on GC cells. We transfected shCAPZA1 plasmid and circSTAU2 overexpression plasmid into GC cells and found that the reduced CAPZA1 expression was rescued by circSTAU2 overexpression (Figure 5A and B). In addition, CAPZA1 knockdown promoted GC cell proliferation, invasion and migration, and these effects were reversed by circSTAU2 overexpression (Figure 5C–F). Taken together, we concluded that circSTAU2 inhibits GC progression by acting as a sponge for miR-589 to remove its suppression on CAPZA1 expression.
MBNL1 Regulates circSTAU2 Expression
It is known that RBP can promote the cyclization of pre-mRNA and plays an important role in the formation of circRNAs.28 To investigate the mechanism of circSTAU2 regulation, we analyzed data from the RBPdb and RBPmap databases and identified two RBPs (Figure 6A). However, the qRT-PCR results showed that only MBNL1 knockdown had an effect on circSTAU2 expression (Figure 6B). To explore the binding site between MBNL1 and STAU2 pre-mRNA, we searched the RBPdb database and identified four potential binding sites (Figure 6C). The results of RIP assay and DNA electrophoresis confirmed that MBNL1 could bind to sites a and d (Figure 6D). Through RNA-FISH and immunofluorescence assays, we found that circSTAU2 and MBNL1 colocalized at the cytoplasm. Besides, MKN45 and BGC823 cells had much lower fluorescence intensities of circSTAU2 and MBNL1 than GES-1 cells (Figure 6E). MBNL1 overexpression enhanced the fluorescence intensity of circSTAU2 and its colocalization at the cytoplasm (Figure 6F). These results showed that MBNL1 promoted the expression of circSTAU2, which acted as a sponge for miR-589 and relieved its suppression on CAPZA1, thus inhibiting the progression of GC.
Exosomes are the Main Carrier of Extracellular circSTAU2
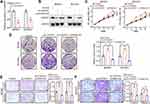
Recent studies have shown that circRNAs are abundant in exosomes.29 CircRNAs play a vital role in tumor progression through exosomes.19–21 To investigate whether extracellular circSTAU2 can inhibit GC progression via exosomes, we examined the existent form of extracellular circSTAU2. The results showed that the circSTAU2 in culture medium was almost equal to the control group when treated with RNase A, but was obviously decreased when treated with RNase A combined with membrane breaker Triton X100 (Figure 7A), suggesting that extracellular circSTAU2 was encapsulated by membrane rather than being released directly. The results of transmission electron microscopy confirmed the existence of exosomes in culture medium of MKN45 and BGC823 cells (Figure 7B). Nano-sight particle tracking analysis showed the number and size distributions of exosomes (Figure 7C). WB analysis showed that exosome marker TSG101 was enriched in exosomes rather than in cell extracts with exosomes removed (Figure 7D). Besides, qRT-PCR results showed that the expression level of circSTAU2 in exosomes was comparable to that in cell culture medium (Figure 7E), indicating that exosomes are the main carrier of extracellular circSTAU2. Further detection of circSTAU2 in exosomes from different cell lines showed that the exosomes from GES-1 cells had the highest abundance, while the exosomes from MKN45 and BGC823 cells had a much lower abundance (Figure 7F).
CircSTAU2 Transmits the Inhibitory Effect on GC Progression by Exosomes
Subsequently, we investigated whether the exosomes encapsulated with circSTAU2 could be taken up by recipient cells. The exosomes extracted from GES-1 cells were labeled with PKH26 dye, and were then incubated with MKN45 and BGC823 cells. The results showed that the exosomes were absorbed by MKN45 and BGC823 cells (Figure 8A). qRT-PCR results showed that the expression of circSTAU2 in recipient cells was significantly increased after co-incubation with exosomes from GES-1 cells, while this effect was weakened when co-incubated with exosomes from circSTAU2-knockdown GES-1 cells (Figure 8B), indicating that exosomes play an important role in the transmission of circSTAU2 between cells. We then explored whether circSTAU2 delivered by exosomes could inhibit GC progression. CCK8 results showed that the growth of MKN45 and BGC823 cells was significantly inhibited after co-incubation with exosomes from GES-1 cells (Figure 8C and D). Interestingly, we found that exosomes from SGC7901 cells could also influence the expression of circSTAU2 and growth of MKN45 and BGC823 cells, although the effects were not that significant as exosomes from GES-1 cells (Supplementary Figure S1A–C). To demonstrate whether exosomes played a key role in this process, we blocked exosome production by the inhibitory effect of neutral sphingomyelinase-2 with GW4869. The results showed that GW4869 significantly reduced the number of exosomes secreted by GES-1 cells (Figure 8E and F). The CCK8 assay showed that co-incubation with the culture medium of GW4869-treated GES-1 cell had no significant inhibitory effect on the growth of MKN45 and BGC823 cells (Figure 8G and H).
In conclusion, we have proved that the inhibitory effect of circSTAU2 on GC progression can be transmitted by exosomes. After taken up by recipient cells, circSTAU2 functions as a sponge for miR-589 to relieve its suppressive effect on CAPZA1, thus inhibiting GC progression (Figure 8I).
Discussion
With the advancement of RNA sequencing technology, an increasing number of circRNAs have been found to be differentially expressed in various cancers, which attracts the great attention of researchers. Recently, numerous studies have proved the crucial role of circRNAs in tumorigenesis.30 For instance, Wei et al found that circASAP1 promoted tumorigenesis and chemoresistance in glioblastoma.31 Yao et al identified that hsa_circ_0058124 facilitated tumorigenesis and invasiveness of papillary thyroid cancer via NOTCH3/GATAD2A axis.32 However, due to the diversity and tissue specificity of circRNAs, the concrete role of circRNAs in GC progression has not been fully elucidated. In our study, we found that circSTAU2 was significantly downregulated in GC. In vitro and in vivo experiments after knockdown or overexpression of circSTAU2 showed that circSTAU2 may act as a tumor inhibitor and had potential use for GC therapy.
Increasing studies reveal that circRNAs can serve as miRNA sponge.33,34 That is, circRNAs function as competing endogenous RNAs to modulate the expression of miRNA target genes by binding to miRNAs.34 For example, circMMP9 accelerates glioblastoma multiforme cell tumorigenesis by targeting miR-124 as a sponge.35 CircCUL2 inhibits GC progression via miR-142-3p/ROCK2 axis.36 In our study, a direct binding between circSTAU2 and miR-589 was predicted and verified. In addition, we found that miR-589 was highly expressed in GC. Overexpressing miR-589 was related to tumor progression, which is consistent with previous studies.37,38
CAPZA1, the α1 subunit of an actin-binding complex that regulates actin cytoskeleton remodeling,27 was confirmed to be a miR-589 target gene. Previous studies showed that CAPZA1 can suppress the metastatic ability of hepatocellular carcinoma cells by inhibiting EMT process,27,39 which is consistent with our results in GC cells. Collectively, our study showed that circSTAU2 inhibits GC progression by the miR-589/CAPZA1 axis. In addition, we explored the mechanism of circSTAU2 regulation and found that MBNL1 plays a role in the upstream of circSTAU2, which binds to STAU2 pre-mRNA and promotes the formation of circSTAU2.
Exosomes, as a new means of information transfer between cells, have been reported to play an important role in tumor progression, which has aroused great interest of researchers.40,41 Recent study shows that exosome-mediated circRNAs are crucial for promoting the progression of colorectal cancer.42 Therefore, after verifying the specific mechanism of circSTAU2 in GC progression, we further explored the role of circSTAU2 in exosomes. As expected, our study showed that circSTAU2 can incorporate into exosomes. After taken up by recipient cells, it can transmit the inhibitory effects on GC progression. This finding suggests that exosomes wrapped with circSTAU2 may be potentially used for GC therapy.
Conclusion
CircSTAU2 is downregulated in GC and circSTAU2 overexpression inhibits GC progression by functioning as a sponge for miR-589 to relieve its inhibitory effect on CAPZA1. Meanwhile, MBNL1 acts as the upstream RBP of circSTAU2 and regulates its expression. Furthermore, we found that circSTAU2 can be incorporated into exosomes and transmit the inhibitory effect on GC progression, which may provide new ideas for GC treatment.
Abbreviations
circRNAs, circular RNAs; GC, gastric cancer; qRT-PCR, quantitative real-time polymerase-chain reaction; WB, Western blot; FISH, fluorescence in situ hybridization; RIP, RNA immunoprecipitation; GEO, gene Expression Omnibus; DECs, differentially expressed circRNAs; TCGA-STAD, The Cancer Genome Atlas Stomach Adenocarcinoma; DEMs, differentially expressed miRNAs; DEGs, differentially expressed genes; RBPs, RNA-binding proteins; HUST, Huazhong University of Science and Technology; FBS, fetal bovine serum; shRNA, short hairpin RNA; WB, Western blot; CCK8, cell counting kit 8; DLR, dual-luciferase reporter; RLUC, renilla luciferase; FLUC, firefly luciferase; PBS, phosphate-buffered saline; SD, standard deviation; EMT, epithelial–mesenchymal transition.
Data Sharing Statement
The datasets analyzed in this study can be found at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). Accession number: GSE93541, GSE184882, GSE64951 and GSE63089.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Union Hospital, Huazhong University of Science and Technology. The approval of animal experiments was granted by the Institutional Animal Care and Use Committee of the Huazhong University of Science and Technology. The mice were managed by professional animal keepers in sterile animal rooms following the recommended procedures of National Institutes of Health guide for the care and use of laboratory animals. All patients signed an informed consent before participating this study.
Acknowledgments
The authors thank Editage for language polishing. Thank Fangqi Chen for the help with figure layout.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by National Natural Science Foundation of China (No. 82073324, 81874184, 82000512) and scientific research project of Hubei health Commission (No. WJ2019Q035).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Moss SF. The clinical evidence linking helicobacter pylori to gastric cancer. Cell Mol Gastroenterol Hepatol. 2017;3(2):183–191. doi:10.1016/j.jcmgh.2016.12.001
3. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–236. doi:10.1136/gutjnl-2017-314205
4. He C, Chen M, Liu J, Yuan Y. Host genetic factors respond to pathogenic step-specific virulence factors of Helicobacter pylori in gastric carcinogenesis. Mutat Res Rev Mutat Res. 2014;759:14–26. doi:10.1016/j.mrrev.2013.09.002
5. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–648. doi:10.1016/S0140-6736(20)31288-5
6. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi:10.15252/embj.2018100836
7. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
8. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
9. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi:10.1038/s41571-021-00585-y
10. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi:10.1186/s12943-020-1135-7
11. Wang X, Xing L, Yang R, et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Mol Cancer. 2021;20(1):91. doi:10.1186/s12943-021-01383-x
12. Wang J, Zhang Y, Song H, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer. 2021;20(1):81. doi:10.1186/s12943-021-01375-x
13. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. doi:10.1007/978-981-13-1426-1_6
14. Chen DL, Sheng H, Zhang DS, et al. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol Cancer. 2021;20(1):166. doi:10.1186/s12943-021-01475-8
15. Shao T, Pan YH, Xiong XD. Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol Ther Nucleic Acids. 2021;23:369–376. doi:10.1016/j.omtn.2020.11.008
16. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi:10.7150/thno.42174
17. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
18. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18(1):75. doi:10.1186/s12943-019-0991-5
19. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
20. Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. doi:10.1186/s12943-020-01189-3
21. Li C, Ni YQ, Xu H, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther. 2021;6(1):383. doi:10.1038/s41392-021-00779-x
22. Wang X, Kang DD, Shen K, et al. An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics. 2012;28(19):2534–2536. doi:10.1093/bioinformatics/bts485
23. Song S, Ajani JA. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2013;10(2):109–118. doi:10.1038/nrgastro.2012.210
24. Ma X, Chen H, Li L, Yang F, Wu C, Tao K. CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition. J Exp Clin Cancer Res. 2021;40(1):330. doi:10.1186/s13046-021-02136-w
25. Du L, Xing Z, Tao B, et al. Both IDO1 and TDO contribute to the malignancy of gliomas via the Kyn-AhR-AQP4 signaling pathway. Signal Transduct Target Ther. 2020;5(1):10. doi:10.1038/s41392-019-0103-4
26. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. CMLS. 2020;77(9):1661–1680. doi:10.1007/s00018-019-03345-5
27. Huang D, Cao L, Zheng S. CAPZA1 modulates EMT by regulating actin cytoskeleton remodelling in hepatocellular carcinoma. CR. 2017;36(1):13. doi:10.1186/s13046-016-0474-0
28. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014
29. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
30. Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57. doi:10.1016/j.canlet.2021.02.004
31. Wei Y, Lu C, Zhou P, et al. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neurooncology. 2021;23(4):611–624. doi:10.1093/neuonc/noaa214
32. Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. CR. 2019;38(1):318. doi:10.1186/s13046-019-1321-x
33. Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16(11):2043–2050. doi:10.1261/rna.2414110
34. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
35. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. doi:10.1186/s12943-018-0911-0
36. Peng L, Sang H, Wei S, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19(1):156. doi:10.1186/s12943-020-01270-x
37. Zhang F, Li K, Pan M, et al. miR-589 promotes gastric cancer aggressiveness by a LIFR-PI3K/AKT-c-Jun regulatory feedback loop. CR. 2018;37(1):152. doi:10.1186/s13046-018-0821-4
38. Zhu H, Zhang H, Pei Y, et al. Long non-coding RNA CCDC183-AS1 acts AS a miR-589-5p sponge to promote the progression of hepatocellular carcinoma through regulating SKP1 expression. CR. 2021;40(1):57. doi:10.1186/s13046-021-01861-6
39. Huang D, Cao L, Xiao L, et al. Hypoxia induces actin cytoskeleton remodeling by regulating the binding of CAPZA1 to F-actin via PIP2 to drive EMT in hepatocellular carcinoma. Cancer Lett. 2019;448:117–127. doi:10.1016/j.canlet.2019.01.042
40. Soltész B, Buglyó G, Németh N, et al. The role of exosomes in cancer progression. Int J Mol Sci. 2021;23(1):1. doi:10.3390/ijms23010008
41. Dai J, Su Y, Zhong S, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi:10.1038/s41392-020-00261-0
42. Shang A, Gu C, Wang W, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19(1):117. doi:10.1186/s12943-020-01235-0
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.