Back to Journals » International Journal of Nanomedicine » Volume 18
Exosomal miR-10527-5p Inhibits Migration, Invasion, Lymphangiogenesis and Lymphatic Metastasis by Affecting Wnt/β-Catenin Signaling via Rab10 in Esophageal Squamous Cell Carcinoma
Authors Xiao Z, Feng X, Zhou Y, Li P, Luo J, Zhang W, Zhou J, Zhao J, Wang D , Wang Y, Tian Z, Zhao X
Received 27 September 2022
Accepted for publication 24 December 2022
Published 6 January 2023 Volume 2023:18 Pages 95—114
DOI https://doi.org/10.2147/IJN.S391173
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yan Shen
Zhaohua Xiao,1 Xumei Feng,2 Yongjia Zhou,1 Peiwei Li,3 Junwen Luo,1 Wenhao Zhang,1 Jie Zhou,1 Jiangfeng Zhao,1 Dong Wang,4 Yongjie Wang,5 Zhongxian Tian,1,6 Xiaogang Zhao1,6
1Department of Thoracic Surgery, The Second Hospital of Shandong University, Jinan, 250033, People’s Republic of China; 2Health Management Center, The Second Hospital of Shandong University, Jinan, People’s Republic of China; 3Institute of Medical Sciences, The Second Hospital of Shandong University, Jinan, People’s Republic of China; 4Department of Thoracic Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, 250021, People’s Republic of China; 5Department of Thoracic Surgery, The Affiliated Hospital of Qingdao University, Qingdao, 266000, People’s Republic of China; 6Key Laboratory of Chest Cancer, Shandong University, The Second Hospital of Shandong University, Jinan, People’s Republic of China
Correspondence: Zhongxian Tian; Xiaogang Zhao, Department of Thoracic Surgery, The Second Hospital of Shandong University, Jinan, 250033, People’s Republic of China, Tel +86-17660082365 ; +86-053185875009, Email [email protected]; [email protected]
Background: Cancer cell-derived exosomal microRNAs (miRNAs) play critical role in orchestrating intercellular communication between tumor cells and tumor microenvironmental factors, including lymphatic endothelial cells (LECs). Nevertheless, the functions and underlying mechanisms of exosomal miRNAs in lymphatic metastasis and lymphangiogenesis in esophageal squamous cell carcinoma (ESCC) remain unclear.
Methods: Small RNA sequencing, Gene Expression Omnibus (GEO) analysis and qRT‒PCR were performed to identify the candidate exosomal miRNAs involved in ESCC metastasis. Receiver operating characteristic curve analysis was conducted to evaluate the diagnostic potential of exosomal miR-10527-5p in predicting lymph node metastasis (LNM) status. An in vitro coculture system was used to investigate the effects of exosomal miR-10527-5p on ESCC cells and human LECs (HLECs), followed by a popliteal LNM assay in vivo. The relationship between miR-10527-5p and Rab10 was identified by dual-luciferase reporter, fluorescence in situ hybridization and qRT‒PCR assays. Then, a series of rescue assays were performed to further investigate whether Rab10 is involved in exosomal miR-10527-5p mediated ESCC metastasis.
Results: MiR-10527-5p was found to be notably reduced in both the plasma exosomes and tumor tissues of ESCC patients with LNM, and plasma exosomal miR-10527-5p had a high sensitivity and specificity for discrimination of LNM status. Moreover, exosome-shuttled miR-10527-5p suppressed the migration, invasion and epithelial-to-mesenchymal transition (EMT) of ESCC cells as well as the migration and tube formation of HLECs via Wnt/β-catenin signaling in vitro and in vivo. Further investigation revealed that Rab10 was a direct target of miR-10527-5p, and re-expression of Rab10 neutralized the inhibitory effects of exosomal miR-10527-5p.
Conclusion: Our study demonstrated that exosomal miR-10527-5p had a strong capability to predict preoperative LNM status and anti-lymphangiogenic effect. Exosomal miR-10527-5p inhibited lymphangiogenesis and lymphatic metastasis of ESCC in a vascular endothelial growth factor-C (VEGF-C)-independent manner, showing potential as a therapeutic target for ESCC patients.
Keywords: esophageal squamous cell carcinoma, exosomes, miR-10527-5p, biomarker, lymphangiogenesis
Introduction
Esophageal carcinoma (EC) is the seventh most frequent malignancy and the sixth leading cause of cancer-related deaths globally, with approximately 572,000 newly diagnosed cases and 509,000 deaths worldwide in 2018.1 There are two main histological types of EC: esophageal adenocarcinoma (EA) and esophageal squamous cell carcinoma (ESCC). ESCC, the predominant subtype in China, comprises approximately 85% of esophageal cancer cases.2 Although the combination of screening and surgical resection has effectively improved the prognosis of early-stage ESCC, the treatment of advanced-stage ESCC is still disappointing.3 As the predominant metastatic method, lymph node metastasis (LNM) is one of the leading causes of the poor prognosis of ESCC patients. Evidence has shown that ESCC patients with LNM (LNM+) have a worse 5-year overall survival (OS) rate than patients without LNM (LNM−).4 Therefore, the identification of a precise molecule that can identify the LNM status and provide insights into the underlying mechanisms of ESCC metastasis is essential for determining treatment strategies and improving the clinical outcomes of ESCC patients.
Exosomes are nanosized lipid bilayer-derived extracellular vesicles ranging in size from 30 to 200 nm.5 Exosomes can be released by multiple types of cells including tumor cells, T cells, B cells and macrophages and serve as mediators of intercellular communication by transmitting information-containing cargos, including proteins, lipids and various types of nucleic acids [including mRNA, microRNA (miRNA), and long noncoding RNA].6,7 The contents of exosomes often vary under different physiological and pathological states and can reflect the state and specific origin of their host cell; as such, they are ideal tools for noninvasive tumor examination.8 Compared with circulating miRNAs, miRNAs in plasma exosomes are protected by the double lipid membrane of exosomes and thus can avoid degradation by RNA enzymes, maintaining their stability. Circulating exosomal miRNAs have also been recognized as promising biomarkers for the early diagnosis, prognostic prediction and recurrence monitoring of ESCC patients.9–11 Nevertheless, whether exosomal miR-10527-5p can be used to determine the LNM status of ESCC patients preoperatively remains unclear and requires further exploration.
Lymphangiogenesis refers to the formation of new lymphatic vessels and has been identified as a new prognostic parameter for predicting the risk of LNM.12 During the sequential processes of lymphatic metastasis, lymphatic vessels in the tumor periphery serve as channels for tumor cell spread to regional lymph nodes (LNs).13 Several cytokines, such as vascular endothelial growth factor C (VEGF-C) and VEGF-D, activate VEGF receptor-3 (VEGFR-3) and induce the proliferation, tube formation and migration of lymphatic endothelial cells (LECs) in the tumor microenvironment.14 Although various inhibitors of the VEGF-C/VEGFR-3 signaling pathway suppressed approximately 60–70% of LNM in a variety of experimental tumor models,15,16 the effectiveness and side effects of these drugs still need to be carefully evaluated. In addition, some ESCC patients with LNM show low levels of VEGF-C. Therefore, exploration of new lymphangiogenesis regulators is important. Recent investigations have demonstrated that exosomes serve as key mediators of lymphangiogenesis in various cancers.17–19 Nevertheless, the roles and regulatory mechanisms of exosomal miR-10527-5p in ESCC lymphangiogenesis remain largely unknown and warrant further investigation.
In the present study, we demonstrated that miR-10527-5p was significantly downregulated in plasma exosomes and tumor tissues of ESCC patients with LNM. In addition, plasma exosomal miR-10527-5p had a strong ability to discriminate LNM status. Moreover, we found that exosomal miR-10527-5p could strikingly inhibit the migration, invasion and epithelial-to-mesenchymal transition (EMT) of ESCC cells as well as the migration and tube formation of human LECs (HLECs), thereby preventing lymphatic metastasis and lymphangiogenesis of ESCC via Wnt/β-catenin signaling by directly targeting Rab10. Our study provides a potential noninvasive biomarker for the preoperative prediction of LNM status in ESCC patients and novel insight into the molecular mechanism of ESCC LNM.
Materials and Methods
Clinical Samples
A total of 62 ESCC patients who underwent surgical resection at the Second Hospital of Shandong University (Jinan, China) from July 2019 to August 2020 were enrolled in this study. All patients underwent esophagectomy with complete resection, and none received any anticancer treatments prior to biopsy collection. Postoperative stage was defined based on the 8th edition of the International Union Against Cancer (UICC) tumor-node-metastasis (TNM) classification criteria. Peripheral blood was collected one week before the operation. During the surgery, tumor tissues were immediately placed into liquid nitrogen.
Small RNA Sequencing and Bioinformatics Analysis
Small RNA sequencing of plasma exosomes from 5 LNM+ and 5 LNM− ESCC patients was performed by Novogene (Beijing, China) as described by Liu.20 Briefly, 3 μg of RNA was used as input material for the generation of a small RNA library. Then, the small RNA library was sequenced on an Illumina HiSeq 2500/2000 platform. Differential expression analysis was performed using the DESeq R package (3.0.3). An adjusted P value of 0.05 and |log2 FC| > 1 were set as the thresholds for significant differential expression. The detailed clinical data of these 10 ESCC patients for small RNA sequencing are presented in Supplementary Table 1. The raw and processed data have been deposited in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; accession number: GSE214259). The GEO dataset GSE155360 contains data from serum exosomes of 20 ESCC patients with or without LNM and was employed to analyze differentially expressed genes by the “limma” package. |log2FC| > 1 was set as the screening threshold. MiRNA target prediction was performed with the miRDB database (mirdb.org), miRWalk database (zmf.umm.uni-heidelberg.de), and miRDIP database (ophid.utoronto.ca).
Cell Culture and Transfection
All cell lines involved in our study except HLECs were purchased from Fu Heng Biological (Shanghai, China). Human ESCC cell lines (KYSE30, Eca109, and KYSE150) were maintained in RPMI-1640 medium (Corning Cellgro, Manassas, VA, USA). Human esophageal epithelial cell lines (HEEC and HET-1A) and HEK293T cells were grown in DMEM (Corning). All media contained 1% penicillin/streptomycin and 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA). HLECs were purchased from ScienCell Research Laboratories (Carlsbad, California, USA) and cultured in endothelial cell medium (ECM; ScienCell) with 5% FBS and 1% endothelial growth medium. All cells were cultured at 37 °C in a 5% CO2 humidified atmosphere.
The miR-10527-5p and Rab10 overexpression lentivirus plasmids were purchased from Genomeditech (Shanghai, China). Puromycin was used at 5 μg/mL for 1 week to select stably transfected cells. The miR-10527-5p mimic, mimic-NC, miR-10527-5p inhibitor and inhibitor-NC were purchased from GenePharma Co., Ltd (Shanghai, China) and transduced into HEK293T or ESCC cells by LipofectamineTM3000 (Invitrogen, Carlsbad, CA, USA). Small interfering RNA (siRNA) against Rab10 (siRab10) and siRNA-NC (si-NC) sequences were synthesized by TSINGKE Biological Technology (Qingdao, China), and these siRNAs were transduced into Eca109 cells by LipofectamineTM RNAiMAX (Invitrogen). The cells were collected after 48–72 h of culture for subsequent experimentation. The primer sequences used are listed in Supplementary Table 2.
Isolation and Identification of Exosomes
For exosome isolation from cell conditioned medium (CM), we first plated an equal number of cells in 10-cm plates, and the cells were rinsed with phosphate-buffered saline (PBS) twice and grown in serum-free medium when they reached 80–90% confluency. After 48 h, the CM was collected and filtered through a 0.22 μm filter (Millipore, Burlington, MA, USA) to remove cells and cell debris, and exosomes were isolated by an exosome isolation kit (System Biosciences, Mountain View, CA, USA). Briefly, the appropriate volume of ExoQuick-TC Exosome Precipitation Solution was added to the CM and refrigerated overnight. The sample was centrifuged at 1500 r.p.m. for 30 min and then at 3000 r.p.m. for 5 min at 4 °C. Finally, the exosome pellet was resuspended in 1×PBS. Additionally, plasma exosomes were isolated using an exosome isolation kit (System Biosciences) according to the manufacturer’s instructions. The exosomes were quantified by bicinchoninic acid (BCA) protein assay and stored at −80 °C for further experiments.
For observation of exosome morphology, exosomes were observed by transmission electron microscopy (TEM, Thermo Scientific, Carlsbad, CA, USA). In addition, exosome-specific protein markers CD63 and TSG101 and the negative marker GRP94 were detected by Western blotting. Moreover, the size distribution of exosomes was measured using NanoSight tracking analysis (NTA) (NanoSight NS300, Malvern, UK) according to the manufacturer’s instructions.
Exosome Labeling and Uptake Assays
Exosomes were labeled with the green fluorescent dye PKH67 (Sigma–Aldrich, MO, USA). Then, the labeled exosomes were incubated with KYSE150 cells or HLECs. After 3 washes with 1×PBS to remove non internalized exosomes, cells were fixed with 4% paraformaldehyde, stained with 4.6-diamidino-2-phenylindole (DAPI; Solarbio, Beijing, China) and analyzed by an Olympus BX43 fluorescence microscope or a confocal microscope (Zeiss, Germany). Additionally, the uptake of exosomes was also assessed by flow cytometry. For visualization of the transport of miR-10527-5p via exosomes, Eca109 cells were transfected with mimic-NC or Cy3-labeled miR-10527-5p mimics. Then, exosomes were isolated from the mimic-NC group or miR-10527-5p mimic group of Eca109 cells, labeled with PKH67, incubated with KYSE150 cells for 24 h, and observed by confocal microscopy.
Quantitative Real-Time PCR (qRT‒PCR)
Total RNA was isolated from cells, exosomes, plasma, and tumor tissues by a miRcute miRNA Isolation Kit (Tiangen Biotech, Beijing, China) and reverse-transcribed into cDNA using a miRNA First-Strand cDNA Synthesis kit or an lnRcute lncRNA First-Strand cDNA synthesis kit (Tiangen). Then, qRT‒PCR was performed using a QuantStudioTM 5 Real-Time PCR System (Thermo Scientific). β-Actin and U6 small nuclear RNA were used as controls for mRNA and miRNA normalization, respectively. The 2−∆∆Ct analytic method was employed to calculate the relative expression levels. The primers used in the current study were synthesized by TSINGKE and are listed in Supplementary Table 3.
Western Blot
Total protein was extracted from the cells or exosomes using RIPA lysis buffer containing protease and phosphatase inhibitors (Beyotime, Shanghai, China). After quantification using a BCA protein assay kit, equal amounts of protein were separated by 10% SDS–PAGE and transferred to PVDF membranes (Millipore). Then, the membranes were blocked in 5% skim milk for 1 h and incubated with primary antibodies at 4 °C overnight. The next day, the membranes were incubated with the corresponding secondary antibodies for 1 h and visualized with an enhanced chemiluminescence (ECL) detection system, and images were captured using a Tanon 5200 system (Tanon, Shanghai, China). GAPDH served as the internal control. The primary antibodies used in this study are shown in Supplementary Table 4.
Immunohistochemistry (IHC)
IHC analysis was performed as we previously described,21 and the results were reviewed and scored separately by two experienced pathologists who were blinded to the clinicopathological data. The immunohistochemical score (IHS) was calculated by combining the proportion of positively stained tumor cells and the staining intensity score. The tumor cell proportion was designated as follows: 0 (no positive tumor cells), 1 (0–10% positive tumor cells), 2 (10–30% positive tumor cells), 3 (30–70% positive tumor cells), and 4 (>75% positive tumor cells). Staining intensity was graded as follows: 1 (no staining), 2 (weak staining), 3 (moderate staining), and 4 (strong staining). Samples with IHS <8 were defined as having low expression, and samples with IHS ≥ 8 were defined as having high expression. The primary antibodies used in the IHC analysis are also shown in Supplementary Table 4.
Wound Healing and Transwell Assays
Briefly, 4×104 cells suspended in 110 µL of medium were added to each well containing a culture insert (Ibidi, Germany) and incubated overnight. When the cells reached 95–100% confluency, the insert that had been scratched was removed to create a wound. After being washed with 1×PBS twice, the cells were cultured with serum-free medium. Images were captured using a Leica DMI8 inverted microscope at 100× magnification every 12 h. The rate of wound healing was analyzed with ImageJ software. The experiment was performed in triplicate.
Uncoated Transwell chambers were used for the migration assay, whereas 50 μL Matrigel (1:10; BD Biosciences, San Jose, CA, USA) was used to precoat the upper surface of the Transwell chambers in the invasion assay. First, 5×104 cells resuspended in 200 μL of serum-free medium were seeded in the upper chamber (8-μm pore size; Corning), and 600 μL of medium with 10% FBS was added in the lower chamber. After incubation for 48 h, the cells that penetrated the lower surface of the membranes were fixed with 4% paraformaldehyde and stained with hematoxylin. Finally, cells from three randomly selected fields were counted. The experiment was performed in triplicate.
Cell Counting Kit-8 (CCK-8), Colony Formation and 5-Ethynyl-2’-Deoxyuridine (EdU) Assays
For the CCK-8 assay, 3×103 cells were plated in 96-well plates. Then, the proliferation level was determined at 0, 24, 48, and 72 h. Ten microliters of CCK-8 kit solution (Dojindo, Tokyo, Japan) with 90 μL of medium was added to each well and then incubated for 2 h. Finally, the optical density (OD) value at 450 nm was measured using a multifunctional enzyme-linked analyzer (BioTek, Winooski, VT, USA).
For the colony formation assay, 1×103 cells were plated in 6-well plates and continuously cultured for approximately 2 weeks. After fixation with 4% paraformaldehyde for 15 min and staining with 0.1% crystal violet for 30 min, visible colonies were imaged and counted.
For the EdU assay, 3×103 cells were seeded in 96-well plates and incubated for 24 h. Briefly, cells were incubated with 100 μL EdU solution (Beyotime) for 2 h and then fixed with 4% paraformaldehyde. After the cells were washed and permeabilized with 3% bovine serum albumin (BSA) and 0.3% Triton X-100, 1× Hoechst 33,342 reaction solution was used to stain nuclei. Finally, images were captured using a fluorescence microscope. The percentage of EdU-positive cells was calculated as follows: (EdU-stained cells/Hoechst-stained cells) ×100%. The experiment was performed in triplicate.
Tube Formation Assay
Matrigel (BD Biosciences) was diluted with precooled culture medium at a ratio of 1:1, placed in 48-well plates (100 μL/well) and polymerized at 37 °C for 1 h. Then, 3×104 HLECs were placed in Matrigel-precoated 48-well plates. After incubation with the corresponding exosomes for 6–8 h, capillary-like structures were photographed under an inverted microscope. The total tube length was calculated using ImageJ software. All experiments were performed in triplicate.
Fluorescence in situ Hybridization (FISH) Assay
FISH was conducted using a RiboTM FISH Kit (RiboBio, Guangzhou, China) as described by Guo.22 Briefly, cells were fixed in 4% paraformaldehyde for 10 min and permeabilized in 0.5% Triton X-100 for 5 min at 4 °C. After incubation with prehybridization buffer at 37 °C for 30 min and hybridization (with a FISH probe) at 37 °C in the dark overnight, the cells were washed with Wash Buffer I (4×SSC), Wash Buffer II (2×SSC), and Wash Buffer III (1×SSC). Then, the cells were incubated with rabbit anti‑Rab10 (dilution 1:200) at 4 °C overnight. The next day, the cells were incubated with Alexa Fluor 488‑conjugated goat anti-rabbit IgG (dilution 1:100) for 1 h at 37 °C, and the nuclei were stained with DAPI in the dark for 10 min. Images were captured using an Olympus BX43 fluorescence microscope (magnification x200). The miR-10527-5p-CY3 FISH probes were designed and synthesized by RiboBio, and the probe sequence was 5’-GCCGTTCACCCAACATTTGCTTT-3’.
RNA Stability Assessment
For evaluation of the effect of miR-10527-5p on the stability of Rab10 mRNA, 20 μM actinomycin D (Sigma–Aldrich) was added to Eca109 cells in the vector group or miR-10527-5p overexpression group and incubated for 0, 1, 2, 3 and 4 h. At the end of the incubation period, total RNA was extracted, and the mRNA level of Rab10 was measured by qRT‒PCR. The experiment was performed in triplicate.
Dual Luciferase Reporter and TOPFlash/FOPFlash Reporter Assay
The 3’ untranslated region (UTR) of Rab10 containing binding sites for miR-10527-5p or its mutated versions were cloned into the pmirGLO vector. HEK293T cells were cotransfected with miR-10527-5p mimic or mimic-NC and luciferase reporter plasmids containing the wild-type (WT) or mutant (MUT) 3’-UTR of Rab10 using LipofectamineTM 3000 reagent (Invitrogen). After transfection for 48 h, luciferase activity was measured by a Dual-Luciferase Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. For the TOPFlash/FOPFlash reporter assay, cells were seeded in 12-well plates and cotransfected with the TOPFlash or FOPFlash and pRL-TK plasmid (Beyotime). After transfection for 48 h, a luciferase assay was performed with the Dual-Luciferase kit. The firefly luciferase activity level was normalized against the Renilla luciferase activity level. The experiment was performed in triplicate.
Popliteal LNM Assay
Female nude mice (BALB/c-nu, 5–6 weeks old) were purchased from HFK Biotechnology (Beijing, China) and were used to construct the popliteal LNM model as described by Liu.23 Briefly, GFP-labeled KYSE150 cells (1×106) with 40 μL of PBS were injected into the footpads of mice. One week after the initial implantation, the nude mice were randomly divided into 2 groups (n = 5/group) and injected intratumorally with Eca109miR-10527-5p-EXOs or Eca109Vector-EXOs (10 μg) twice a week. Lymphatic metastasis was monitored and imaged with an In Vivo Imaging System (Spectrum CT, PerkinElmer, USA). Four weeks after the initial implantation, the footpad tumors and popliteal LNs were excised, measured (volume = width2 × length/2) and paraffin embedded for further analysis. Positive LNs were indicated by GFP-positive tumor cells detected by IHC. The ratios of positive-stained LNs to total LNs were calculated.
Statistical Analysis
Statistical analyses were conducted with GraphPad Prism 8.0.1 (GraphPad, San Diego, CA, USA). Differences between groups were analyzed by Student’s t-test for comparisons between two groups or one-way analysis of variance (ANOVA) for multiple comparisons. The chi-square test was applied for categorical variables. The correlation of measurements was determined using Pearson’s correlation analysis. Measurement data are presented as the mean ± standard deviation (SD), and P < 0.05 was considered significant.
Results
MiR-10527-5p is Reduced in Plasma Exosomes and Tumor Tissues of ESCC Patients with LNM
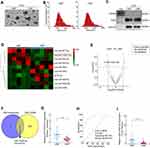
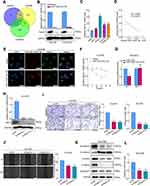
Initially, exosomes were isolated from plasma and tissues from LNM+ ESCC or LNM− ESCC patients by an ExoQuick exosome isolation kit. Then, TEM was performed to verify the typical disc-shaped morphology of the isolated exosomes (EXOs, Figure 1A). NTA showed that the median diameters of plasma exosomes were 128.1 (LNM+) and 126.0 nm (LNM−) (Figure 1B). In addition, Western blotting confirmed the expression of the exosome markers CD63 and TSG101 and the negative marker GRP94 (Figure 1C). With the goal of identifying candidate exosomal miRNAs involved in ESCC metastasis, we conducted small RNA sequencing to determine the miRNA expression profiles of plasma exosomes from 5 LNM+ and 5 LNM− ESCC patients. As depicted in Figure 1D and E, we identified 10 exosomal miRNAs with |log2FC|> 1 and P<0.05 (Supplementary Table 5), 4 miRNAs that were significantly downregulated and 6 miRNAs that were significantly upregulated in the plasma exosomes of LNM+ patients compared to LNM− patients. We further analyzed the GEO dataset GSE155360, which contains the miRNA expression datasets of serum exosomes from 20 ESCC patients, and used it in combined analysis with our sequencing data. As a result, only miR-10527-5p and miR-493-5p were found to be differentially expressed in the RNA sequencing data and GSE155360 dataset (Figure 1F). Of these, miR-10527-5p (a product of the MIR10527 gene, which is located on chromosome 12q14.2) showed a smaller P value and was selected for further experiments. Then, qRT‒PCR was applied to validate the expression of miR-10527-5p in plasma exosomes of 52 ESCC patients (26 LNM− and 26 LNM+ ESCC patients). As shown in Figure 1G, exosomal miR-10527-5p expression was significantly lower in the LNM+ group than in the LNM− group, which was consistent with the results of RNA sequencing. In addition, receiver operating characteristic (ROC) curve analysis showed that plasma exosomal miR-10527-5p had a high accuracy for distinguishing LNM+ patients from LNM− patients. The area under the ROC curve (AUC) value, sensitivity and specificity were 0.8935, 84.42% and 80.75%, respectively (Figure 1H), highlighting the diagnostic potential of plasma exosomal miR-10527-5p in predicting the LNM status of ESCC patients. We also evaluated the miR-10527-5p expression levels in ESCC tumor tissues by qRT‒PCR. Consistent with the expression of miR-10527-5p in plasma exosomes, miR-10527-5p was substantially reduced in tumor tissues of LNM+ patients (n=26) compared with LNM− patients (n=26) (Figure 1I). Taken together, these data indicated that miR-10527-5p plays a crucial role in the progression of ESCC metastasis.
Downregulation of Exosomal MiR-10527-5p is Associated with Poor Clinicopathological Characteristics of ESCC Patients
Fifty-two ESCC patients were classified into 2 groups based on the median miR-10527-5p levels in plasma exosomes, with 26 patients in the low-expression group and 26 patients in the high-expression group. Then, the chi-square test was used to investigate the association between exosomal miR-10527-5p levels and clinicopathological parameters (including sex, age, tumor size, T stage, differentiation degree, and LNM status). As shown in Table 1, exosomal miR-10527-5p levels were significantly negatively correlated with tumor size (P=0.002), T stage (P=0.004) and LNM status (P =0.012), while no significant associations were observed between exosomal miR-10527-5p expression and sex, age or differentiation degree (P>0.05). These data indicated that downregulation of exosomal miR-10527-5p was strongly associated with poor clinicopathological characteristics of ESCC patients.
 |
Table 1 Correlations Between Plasma Exosomal miR-10527-5p Expression and Clinicopathologic Characteristics of 52 ESCC Patients |
MiR-10527-5p Inhibits the Migration, Invasion and EMT of ESCC Cells via Wnt/β-Catenin Signaling
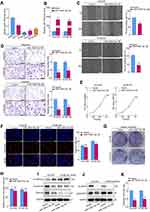
Based on the above data, we further assessed whether miR-10527-5p mediates the biological functions of ESCC cells. First, we compared the expression levels of miR-10527-5p in three human ESCC cell lines (KYSE30, Eca109, and KYSE150) and two esophageal epithelial cell lines (HET-1A and HEEC) via qRT‒PCR. As shown in Figure 2A, miR-10527-5p expression was considerably lower in the ESCC cell lines than in the normal esophageal epithelial cell lines. Then, Eca109 and KYSE150 cells were transfected with miR-10527-5p overexpression (miR-10527-5p OE) lentiviruses, and qRT‒PCR was used to confirm the transfection efficiency (Figure 2B). Wound healing and Transwell assays demonstrated that miR-10527-5p OE significantly suppressed the migration and invasion of Eca109 and KYSE150 cells (Figure 2C and D). Conversely, the miR-10527-5p inhibitor (inh-miR-10527-5p, Figure S1A) dramatically enhanced the cell migration and invasion of Eca109, KYSE150 and KYSE30 cells (Figure S1B–S1E). However, there was no obvious significant difference in cell proliferation as determined by CCK-8, EdU and clone formation assays (Figure 2E–H). EMT, an evolutionarily conserved developmental program, has been implicated in carcinogenesis and confers metastatic properties on cancer cells by enhancing cell mobility, migration and invasive potential.24 As shown in Figure 2I, the expression of mesenchymal markers (N-cadherin and Vimentin) was significantly reduced whereas that of the epithelial marker E-cadherin was increased after miR-10527-5p overexpression. However, miR-10527-5p inhibitor treatment resulted in the opposite effects (Figure S1F). Accumulating evidence has revealed that Wnt/β-catenin signaling is involved in EMT and tumor metastasis.25 Therefore, we next explored whether miR-10527-5p regulation of ESCC EMT could be mediated by the Wnt/β-catenin pathway. As shown in Figure 2J, phosphorylated GSK-3βSer9 (pGSK3βSer9) and β-catenin levels were significantly reduced after miR-10527-5p overexpression but increased after administration of the miR-10527-5p inhibitor (Figure S1G). Then, TOPFlash and FOPFlash reporters, containing WT and MUT β-catenin/TCF-4 binding sites respectively, were constructed to verify whether miR-10527-5p affects the transcriptional activity of β-catenin. Notably, overexpression of miR-10527-5p strongly decreased the activity of TOP/FOP (Figure 2K). However, TOP/FOP activity was increased when miR-10527-5p was downregulated (Figure S1H). These data suggest that miR-10527-5p inhibits the migration, invasion and EMT of ESCC cells through the Wnt/β-catenin pathway.
Exosomal MiR-10527-5p Inhibits the Migration, Invasion and EMT of ESCC Cells via Wnt/β-Catenin Signaling
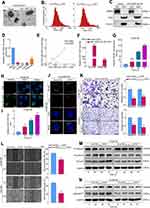
To investigate whether miR-10527-5p is enriched by ESCC-derived exosomes, we purified exosomes from the CM of Eca109 cells and then identified them by TEM, NTA, and Western blot assays (Figure 3A–C). qRT‒PCR results revealed that miR-10527-5p levels were significantly lower in ESCC cell-secreted exosomes than in exosomes isolated from human esophageal epithelial cells (Figure 3D), and the levels of exosomal miR-10527-5p were significantly positively correlated with the levels in cells (R=0.899, P=0.038, Figure 3E). In addition, miR-10527-5p was more highly expressed in exosomes extracted from miR-10527-5p-overexpressing cells (miR-10527-5p OE-EXOs) than in those extracted from cells transfected with the control vector (Vector-EXOs) (Figure 3F). During tumor metastasis, tumor-derived exosomal miRNAs can be taken up by other tumor cells and regulate their biological behaviors. Eca109 cells that showed low levels of E-cadherin and high levels of Vimentin and N-cadherin were designated high-EMT cells, and KYSE150 cells that expressed high levels of E-cadherin and no detectable levels of Vimentin or N-cadherin were designated low-EMT cells. Furthermore, an in vitro coculture system was used to investigate the functional roles of exosomal miR-10527-5p in intercellular communication between ESCC cells. PKH67-labeled exosomes derived from Eca109 cells were cocultured with KYSE150 cells. Flow cytometry results demonstrated that the PKH67 fluorescence intensity in KYSE150 cells increased with time (Figures 3G and S2A). Interestingly, we observed that the green fluorescent signal colocalized mainly in the perinuclear region of KYSE150 cells using fluorescence microscopy (Figures 3H and S2B). The qRT‒PCR results revealed that Eca109miR-10527-5p-EXOs obviously increased the level of miR-10527-5p in KYSE150 cells in a time-dependent manner (Figure 3I). Additionally, in KYSE150 cells, colocalization of Cy3-labeled miR-10527-5p mimics and PKH67 fluorescence was observed in the Eca109miR-10527-5p-EXO group (Figure 3J). Taken together, these results indicated that horizontal transfer of miR-10527-5p from Eca109 cells to KYSE150 cells can occur via exosomes. Wound healing and Transwell assays demonstrated that Eca109miR-10527-5p-EXOs could significantly decrease the cell migratory and invasive capacities of KYSE150 and Eca109 cells (Figure 3K–L). Interestingly, after incubation with Eca109Vector-EXOs for 0, 24, 48 and 72 h, the expression levels of Vimentin, N-cadherin, pGSK3βSer9 and β-catenin in KYSE150 cells were significantly increased, and that of E-cadherin was decreased, whereas there were no significant changes after incubation with Eca109miR-10527-5p-EXOs (Figure 3M and N), which indicated that exosomal miR-10527-5p inhibits the migration, invasion and EMT of ESCC cells through the Wnt/β-catenin pathway.
Exosomal MiR-10527-5p Inhibits Lymphatic Metastasis and Lymphangiogenesis in vitro and in vivo
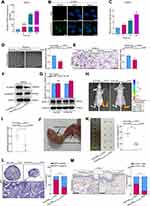
To investigate whether exosomal miR-10527-5p mediates the migration and tube formation of HLECs, we treated HLECs with different exosomes derived from Eca109 cells. The exosome uptake assay (Figure 4A and B and Figure S2C) and qRT‒PCR (Figure 4C) results showed that exosomal miR-10527-5p could also be transported from Eca109 cells to HLECs. Subsequently, tube formation and Transwell assays were performed to assess the impacts of exosomal miR-10527-5p on HLECs. We observed that Eca109miR-10527-5p-EXOs notably inhibited HLEC tube formation and migration compared with Eca109Vector-EXOs (Figure 4D and E). Moreover, the Western blot results demonstrated that pGSK3βser9 and β-catenin levels significantly decreased when HLECs were incubated with Eca109miR-10527-5p-EXOs for 72 h (Figure 4F), suggesting that exosomal miR-10527-5p inhibits the migration and tube formation of HLECs via the Wnt/β-catenin pathway. As a classic lymphangiogenic growth factor, VEGF-C is a key regulator of lymphangiogenesis. Thus, the relationship between miR-10527-5p and VEGF-C expression was further examined. The results revealed that miR-10527-5p overexpression did not influence VEGF-C mRNA and protein levels (Figure 4G). Collectively, these findings indicated that exosomal miR-10527-5p inhibits the migration and tube formation of HLECs in a VEGF-C-independent manner. The regulatory function of exosomal miR-10527-5p in ESCC lymphangiogenesis was evaluated in vivo. An in vivo imaging system (IVIS) revealed that Eca109miR-10527-5p-EXOs strikingly prohibited the metastasis of the primary tumor to the popliteal LNs compared with Eca109Vector-EXOs (Figure 4H and I). Four weeks after the initial implantation, foot-pad tumors and popliteal LNs were enucleated (Figure 4J). Strikingly, Eca109miR-10527-5p-EXO injection obviously reduced the volume of popliteal LNs compared with Eca109Vector-EXO injection (Figure 4K). Similarly, the Eca109miR-10527-5p-EXO group showed a lower rate of metastasis-positive popliteal LNs, as indicated by GFP-positive tumor cells detected by IHC (Figure 4L). Additionally, IHC analysis revealed that the peritumoral lymphatic vessel density (LVD) in footpad tumor tissues was significantly lower in the Eca109miR-10527-5p-EXO group than in the Eca109Vector-EXO group (Figure 4M). Taken together, these results indicated that exosomal miR-10527-5p inhibits lymphatic metastasis and lymphangiogenesis of ESCC in vitro and in vivo.
Rab10 is a Direct Target Gene of miR-10527-5p
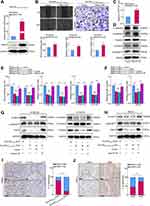
To investigate the factor through which miR-10527-5p mediates ESCC cell metastasis, we used three bioinformatics databases (miRWalk, miRDB, and miRDIP) to identify the candidate target genes of miR-10527-5p (Figure 5A). As shown in Figures 5B and S3A, Ras-related protein Rab-10 (Rab10) showed the most significant decrease in mRNA levels among the 5 candidate target genes after miR-10527-5p overexpression. Consistently, the Western blot results showed that the Rab10 protein level was also substantially suppressed (Figure 5B). Then, we compared the mRNA levels of Rab10 in KYSE30, Eca109, KYSE150, HET-1A and HEEC cells via qRT‒PCR. As shown in Figure 5C, Rab10 expression was considerably higher in the ESCC cell lines than in HET-1A and HEEC cells, and the level of Rab10 tended to negatively correlate with the miR-10527-5p level (R=0.844, P=0.072, Figure 5D). In addition, the FISH assay showed a negative correlation between the expression of miR-10527-5p and Rab10, which colocalized mainly in the cytoplasm of Eca109 cells (Figure 5E). Evidence has shown that miRNAs can modulate gene expression by directly targeting mRNAs, resulting in their post-transcriptional suppression or instability.26 In the present study, we found that Rab10 was more stable in the miR-10527-5p overexpression group than in the vector group under treatment with actinomycin D (Figure 5F), which indicated that miR-10527-5p decreased Rab10 through post-transcriptional suppression rather than destabilization. To examine whether miR-10527-5p directly targets Rab10 mRNA, we performed dual luciferase reporter analysis in HEK293T cells. We found that the miR-10527-5p mimic effectively inhibited the activity of the firefly luciferase reporter containing the WT 3’-UTR of Rab10 compared with the mimic-NC, whereas this effect was abolished when the binding sites were mutated (Figure 5G), which indicated that Rab10 is a direct target of miR-10527-5p. The miR-10527-5p binding site in the Rab10 mRNA 3’-UTR is shown in Figure S3B. To verify the biological functions of Rab10 in ESCC cells, we downregulated Rab10 by siRNA (siRab10, Figure 5H). In vitro studies revealed that silencing Rab10 dramatically attenuated the migration and invasion of Eca109 cells (Figure 5I and J). Moreover, knockdown of Rab10 notably inactivated Wnt/β-catenin signaling and reversed EMT progression in Eca109 cells (Figure 5K and L).
Exosomal MiR-10527-5p Targets Rab10 to Inhibit ESCC Metastasis
To determine whether Rab10 was necessary for miR-10527-5p-mediated ESCC metastasis, we transfected Eca109miR-10527-5p OE cells with Rab10 overexpression lentiviruses (Rab10 OE, Figure 6A). Wound healing and Transwell assays revealed that upregulation of Rab10 significantly reversed the suppression of Eca109 cell migration and invasion due to miR-10527-5p OE (Figure 6B). The results of the TOPFlash/FOPFlash reporter assay and Western blots showed that the inhibitory effects of miR-10527-5p OE on Wnt/β-catenin signaling and EMT progression were also reversed when Rab10 was reintroduced (Figure 6C and D). To further investigate whether Rab10 is involved in exosomal miR-10527-5p-mediated lymphatic metastasis and lymphangiogenesis in ESCC, we performed a series of rescue assays. In vitro studies revealed that cotreatment with Eca109miR-10527-5p-EXOs and the Rab10 overexpression plasmid significantly reversed the inhibitory effects of exosomal miR-10527-5p on the migration and invasion of KYSE150 and Eca109 cells (Figures 6E and S4A). Similarly, re-expression of Rab10 also neutralized exosomal miR-10527-5p-mediated inhibition of the migration and tube formation of HLECs (Figures 6F and S4B). Likewise, the Western blot results showed that the inhibitory effects of exosomal miR-10527-5p on Wnt/β-catenin signaling and EMT in KYSE150 cells (Figure 6G), as well as Wnt/β-catenin signaling in HLECs (Figure 6H), were also significantly reversed when Rab10 was reintroduced. Then, we detected Rab10 expression levels in footpad tumor tissues by IHC. As shown in Figure 6I, Rab10 expression was significantly lower in the Eca109miR-10527-5p-EXO group than in the Eca109Vector-EXO group. Based on the Kaplan–Meier plotter online database,27 we generated a Kaplan–Meier OS curve, which revealed that the OS rate was obviously reduced in ESCC patients with Rab10 overexpression (Figure S5). In addition, IHC assays indicated that Rab10 expression in LNM+ ESCC tumor tissues was significantly higher than that in LNM− specimens (Figure 6J). Collectively, these data indicate that exosomal miR-10527-5p inhibits lymphatic metastasis and lymphangiogenesis in ESCC by directly targeting Rab10 (Figure 7).
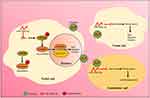 |
Figure 7 Proposed model of exosomal miR-10527-5p-mediated migration, invasion, and EMT of ESCC cells and lymphangiogenesis of HLECs. |
Discussion
In the current study, we identified an exosomal miRNA, miR-10527-5p, that was reduced in the plasma exosomes of ESCC patients with LNM. In addition, plasma exosomal miR-10527-5p showed a strong ability to discriminate the preoperative LNM status. Moreover, we found that exosomal miR-10527-5p could significantly attenuate the migration, invasion and EMT of ESCC cells by blocking Rab10-mediated Wnt/β-catenin signaling. Furthermore, exosomal miR-10527-5p could be transferred to HLECs, in which it exerted its anti-lymphangiogenic effects in a VEGF-C-independent manner. Our study provides a noninvasive biomarker for predicting the LNM status of ESCC patients and novel insight into the molecular mechanism of ESCC metastasis.
A key initial finding made during the present study was the clinical value of plasma exosomal miR-10527-5p in predicting preoperative LNM status. Currently, imaging technologies such as computed tomography (CT) and endoscopic ultrasonography (EUS) are the main methods used by clinicians to determine the N stage of ESCC patients. However, due to the low specificity and sensitivity of CT and EUS, in some cases, micrometastases are not detected, which results in incorrect staging and inadequate therapy.28,29 Recently, exosomal miRNAs have attracted extensive attention for their clinical utility in predicting LNM status.30,31 Compared with CT and EUS, the detection of exosomal miRNAs is less traumatic and more convenient. In the current study, we found that plasma exosomal miR-10527-5p had a high sensitivity and specificity in determining the LNM status of ESCC patients, suggesting that exosomal miR-10527-5p is a promising biomarker for the selection of patients with a high risk of LNM. Additionally, the plasma exosomal miR-10527-5p levels were negatively correlated with the LNM status of ESCC patients, highlighting the essential role of miR-10527-5p in ESCC metastasis.
Another important finding in the current study was the anti-lymphangiogenic effect of exosomal miR-10527-5p. Several studies have demonstrated that exosomal miRNAs serve as key mediators of tumor lymphangiogenesis. Nevertheless, the functions and molecular mechanisms of exosomal miRNAs in ESCC lymphangiogenesis remain unclear. In vitro studies, our results revealed that Eca109-secreted exosomal miR-10527-5p could be transferred to HLECs to inhibit their migration and tube formation. The anti-lymphangiogenic effect of exosomal miR-10527-5p was further confirmed by a popliteal LN metastasis assay. Above all, a decrease in exosomal miR-10527-5p might be the main contributor to ESCC microenvironment generation. Conversely, a recent study revealed that exosomal miR-320b promotes lymphangiogenesis and lymphatic metastasis of ESCC by targeting PDCD4.32 In addition, García-Silva found that melanoma-derived exosomes could induce lymphangiogenesis and metastasis through nerve growth factor receptor (NGFR).33 All of these indicate that whether exosomes are pro-lymphangiogenic or anti-lymphangiogenic probably depends on the tissue type and the distinct cargo composition of exosomes. Tumor lymphangiogenesis can be modulated by multiple regulators, such as VEGF-C. However, there were no obvious changes in the mRNA and protein levels of VEGF-C after miR-10527-5p overexpression. Growing evidence suggests essential roles for the Wnt/β-catenin signaling pathway in lymphangiogenesis.34,35 Given the results of published literature and our previous results, we hypothesized that the Wnt/β-catenin signaling pathway might contribute to the inhibitory effects of exosomal miR-10527-5p on lymphangiogenesis. The central event in the activation of Wnt/β-catenin signaling is the accumulation of β-catenin in the cytoplasm, which is followed by the translocation of unphosphorylated β-catenin to the nucleus, where it forms a β-catenin-TCF/LEF transcriptional complex and induces the transcription of target genes. GSK3β mediates the phosphorylation of β-catenin, promoting its ubiquitination and subsequent proteasomal degradation, whereas phosphorylation and inhibition of GSK3β ensure an increase in cytosolic β-catenin concentration.36,37 To verify the above assumption, we performed Western blotting to detect the expression of pGSK3βSer9 and β-catenin in HLECs. The results showed that exosomal miR-10527-5p significantly decreased the expression of pGSK3βSer9 and β-catenin, which confirmed that the Wnt/β-catenin signaling pathway plays a crucial role in lymphangiogenesis in ESCC.
In addition, we investigated the functional role of exosomal miR-10527-5p in intercellular communication between ESCC cells. A previous study identified different cell subpopulations within the same tumor, and the gene expression and migration and invasion differ among different subpopulations.38 During tumor metastasis, tumor-derived exosomal miRNAs can be internalized by other tumor cells, thereby affecting recipient cell activities, such as proliferation, invasion, EMT, angiogenesis, and drug resistance.39 Liu et al reported that exosomes from colorectal cancer (CRC) cells with high invasive potential can deliver miR-106b-3p to CRC cells with low invasive potential, which enhances CRC cell migration and invasion by targeting DLC-1.20 Our results showed that miR-10527-5p could not only inhibit the migration, invasion and EMT of ESCC cells but also be horizontally delivered to KYSE150 cells from Eca109 cells via exosomes, thus inhibiting their migration, invasion and EMT. Based on these results, we proved that exosomal miR-10527-5p has a major regulatory effect on the metastatic potential of ESCC cells.
Further investigation demonstrated that Rab10 is a direct target gene of miR-10527-5p. Rab10, a member of the small GTPase family,40 has been identified as an oncogene in various cancers, such as breast cancer, osteosarcoma, and ESCC, and it participates in tumor metastasis by affecting different processes, including invasion, migration, EMT, immune evasion and chemoresistance.41–43 In our study, Rab10 was found to be significantly elevated in LNM+ ESCC tissues and tended to be associated with poor prognosis in ESCC patients. Moreover, we verified that silencing Rab10 decreased the expression of pGSK3βSer9 and β-catenin and inhibited the migration, invasion, and EMT of ESCC cells. However, upregulation of Rab10 reversed the inhibitory effects of miR-10527-5p overexpression. Most importantly, re-expression of Rab10 obviously neutralized exosomal miR-10527-5p-mediated inhibition of the migration, invasion and EMT of ESCC cells, as well as exosomal miR-10527-5p-mediated inhibition of the migration and tube formation of HLECs. All of these results demonstrated that exosomal miR-10527-5p inhibits lymphatic metastasis and lymphangiogenesis by directly targeting Rab10, and targeting Rab10 is promising strategy for ESCC patients with LNM. Given the low efficacy and side effects of current treatment approaches, which include chemotherapy, radiotherapy, immunotherapy and targeted therapy, it is desirable to develop novel therapeutic strategies for advanced-stage ESCC patients. Recent investigations have demonstrated that exosome drug delivery systems have great application prospects in tumor therapy owing to their high stability, low cytotoxicity and minimal immunogenicity.44,45 Therefore, miR-10527-5p packaging into exosomes may overcome the shortcomings of current treatment methods and serve as a potential therapeutic strategy for ESCC patients. There are several limitations in the present study. First, the number of samples was relatively small, which may affect the reliability of our findings. Therefore, larger sample sizes are required to verify our results. Second, the prognostic value of Rab10 as well as the detailed mechanism by which Rab10 regulates the Wnt/β-catenin pathway remains unclear and requires further investigation. Third, in this study, we mainly focused on the role and mechanism of tumor-derived exosomal miR-10527-5p, and whether miR-10527-5p could be carried by exosomes from immune cells or other stromal cells also needs to be investigated in future experiments.
Conclusion
In conclusion, our study first verified that plasma exosomal miR-10527-5p levels were negatively correlated with lymphatic metastasis in ESCC patients and that plasma exosomal miR-10527-5p might serve as a noninvasive biomarker for predicting LNM status. Exosomal miR-10527-5p could be transferred to ESCC cells and HLECs and suppressed the migration, invasion and EMT of ESCC cells as well as the migration and tube formation of HLECs, thereby preventing lymphatic metastasis and lymphangiogenesis of ESCC through the Wnt/β-catenin signaling pathway by directly targeting Rab10. Therefore, exosomal miR-10527-5p could serve as a diagnostic biomarker and potential therapeutic target for ESCC treatment.
Abbreviations
ESCC, esophageal squamous cell carcinoma; LNM, lymph node metastasis; LNs, lymph nodes; VEGF, vascular endothelial growth factor; HLECs, human lymphatic endothelial cells; CM, conditioned medium; FISH, fluorescence in situ hybridization; EXO, exosomes; EMT, epithelial-to-mesenchymal transition; pGSK3βSer9, phosphorylated GSK-3βSer9; miR-10527-5p OE-EXOs, exosomes extracted from miR-10527-5p-overexpressing cells; Vector-EXOs, exosomes extracted from cells transfected with control vector; ROC, receiver operating characteristic; AUC, area under the ROC curve; LVD, lymphatic vessel density.
Data Sharing Statement
All data generated for this study are included in the article and its Supplementary Materials Files.
Ethics Statement
This study was conducted according to the principles expressed in the Declaration of Helsinki and approved by the Ethics Committee of the Second Hospital of Shandong University (protocol code: KYLL-2020(LW)-082). Written informed consent was obtained from each patient.
Acknowledgments
The authors expressed their gratitude to all donors for sample donations and the American Journal Experts for providing language editing.
Funding
This study was funded by the grant from the Special Construction Project Fund for Taishan Mountain Scholars of Shandong Province, Jinan Clinical Medicine Research Program for Thoracic Cancer, and Natural Science Foundation of Shandong Province of China (grant number. ZR2019MH092).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Arnold M, Soerjomataram I, Ferlay J, et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi:10.1136/gutjnl-2014-308124
3. Thrift A. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nature Rev Gastroenterol Hepatol. 2021;18(6):432–443. doi:10.1038/s41575-021-00419-3
4. Liu Y, Ma L, Wang S, et al. Prognostic value of lymph node metastases and lymph node ratio in esophageal squamous cell carcinoma. Eur J Surg Oncol. 2010;36(2):155–159. doi:10.1016/j.ejso.2009.09.005
5. Pegtel D, Gould SE. Exosomes. Annual Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
6. van den Boorn J, Dassler J, Coch C, et al. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65(3):331–335. doi:10.1016/j.addr.2012.06.011
7. Shen M, Ren X. New insights into the biological impacts of immune cell-derived exosomes within the tumor environment. Cancer Lett. 2018;431:115–122. doi:10.1016/j.canlet.2018.05.040
8. Yu W, Hurley J, Roberts D, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Annals Oncol. 2021;32(4):466–477. doi:10.1016/j.annonc.2021.01.074
9. Liu S, Lin Z, Zheng Z, et al. Serum exosomal microRNA-766-3p expression is associated with poor prognosis of esophageal squamous cell carcinoma. Cancer Sci. 2020;111(10):3881–3892. doi:10.1111/cas.14550
10. Jing Z, Chen K, Gong L. The significance of exosomes in pathogenesis, diagnosis, and treatment of esophageal cancer. Int J Nanomedicine. 2021;16:6115–6127. doi:10.2147/IJN.S321555
11. Li K, Lin Y, Luo Y, et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer. 2022;21(1):21. doi:10.1186/s12943-022-01499-8
12. Stacker S, Williams S, Karnezis T, et al. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nature Rev Cancer. 2014;14(3):159–172. doi:10.1038/nrc3677
13. Sundar S, Ganesan T. Role of lymphangiogenesis in cancer. J Clin Oncol. 2007;25(27):4298–4307. doi:10.1200/JCO.2006.07.1092
14. Hirakawa S, Brown L, Kodama S, et al. VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood. 2007;109(3):1010–1017. doi:10.1182/blood-2006-05-021758
15. Burton J, Priceman S, Sung J, et al. Suppression of prostate cancer nodal and systemic metastasis by blockade of the lymphangiogenic axis. Cancer Res. 2008;68(19):7828–7837. doi:10.1158/0008-5472.CAN-08-1488
16. Hwang S, Song J, Kim Y, et al. Inhibition of lymphatic proliferation by the selective VEGFR-3 inhibitor SAR131675 ameliorates diabetic nephropathy in db/db mice. Cell Death Dis. 2019;10(3):219. doi:10.1038/s41419-019-1436-1
17. Zhou CF, Ma J, Huang L, et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene. 2019;38(8):1256–1268. doi:10.1038/s41388-018-0511-x
18. Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421. doi:10.1172/JCI130892
19. Zheng H, Chen C, Luo Y, et al. Tumor-derived exosomal BCYRN1 activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clin Transl Med. 2021;11(7):e497. doi:10.1002/ctm2.497
20. Liu H, Liu Y, Sun P, et al. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci. 2020;134(4):419–434. doi:10.1042/CS20191087
21. Xiao Z, Tian Y, Jia Y, et al. RUNX3 inhibits the invasion and migration of esophageal squamous cell carcinoma by reversing the epithelial‑mesenchymal transition through TGF‑β/Smad signaling. Oncol Rep. 2020;43(4):1289–1299. doi:10.3892/or.2020.7508
22. Guo X, Xu Y, Wang Z, et al. A Linc1405/eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell. 2018;22(6):893–908.e896. doi:10.1016/j.stem.2018.04.013
23. Liu L, Lin C, Liang W, et al. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut. 2015;64(1):26–36. doi:10.1136/gutjnl-2013-306388
24. Pastushenko I, Blanpain C, Transition EMT. States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
25. Ghahhari N, Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Euro J Cancer. 2015;51(12):1638–1649. doi:10.1016/j.ejca.2015.04.021
26. Ni W, Leng X. miRNA-dependent activation of mRNA translation. MicroRNA. 2016;5(2):83–86.
27. Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Inter Res. 2021;23(7):e27633. doi:10.2196/27633
28. Thosani N, Singh H, Kapadia A, et al. Diagnostic accuracy of EUS in differentiating mucosal versus submucosal invasion of superficial esophageal cancers: a systematic review and meta-analysis. Gastrointest Endosc. 2012;75(2):242–253. doi:10.1016/j.gie.2011.09.016
29. Jeong D, Kim M, Lee K, et al. Surgically resected T1- and T2-stage esophageal squamous cell carcinoma: t and N staging performance of EUS and PET/CT. Cancer Med. 2018;7(8):3561–3570. doi:10.1002/cam4.1617
30. Jiang K, Li G, Chen W, et al. Plasma Exosomal miR-146b-5p and miR-222-3p are potential biomarkers for lymph node metastasis in papillary thyroid carcinomas. Onco Targets Ther. 2020;13:1311–1319. doi:10.2147/OTT.S231361
31. Wang X, Qian T, Bao S, et al. Circulating exosomal miR-363-5p inhibits lymph node metastasis by downregulating PDGFB and serves as a potential noninvasive biomarker for breast cancer. Mol Oncol. 2021;15(9):2466–2479. doi:10.1002/1878-0261.13029
32. Liu T, Li P, Li J, et al. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol Ther Oncolytics. 2021;23:163–180. doi:10.1016/j.omto.2021.09.003
33. García-Silva S, Benito-Martín A, Nogués L, et al. Melanoma-derived small extracellular vesicles induce lymphangiogenesis and metastasis through an NGFR-dependent mechanism. Nature Cancer. 2021;2(12):1387–1405. doi:10.1038/s43018-021-00272-y
34. Stump B, Shrestha S, Lamattina A, et al. Glycogen synthase kinase 3-β inhibition induces lymphangiogenesis through β-catenin-dependent and mTOR-independent pathways. PLoS One. 2019;14(4):e0213831. doi:10.1371/journal.pone.0213831
35. Wang S, Chang J, Hsiao J, et al. Tumour cell-derived WNT5B modulates in vitro lymphangiogenesis via induction of partial endothelial-mesenchymal transition of lymphatic endothelial cells. Oncogene. 2017;36(11):1503–1515. doi:10.1038/onc.2016.317
36. Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13(1):165. doi:10.1186/s13045-020-00990-3
37. Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi:10.1016/j.cell.2017.05.016
38. Lawson D, Kessenbrock K, Davis R, et al. Tumour heterogeneity and metastasis at single-cell resolution. Nature Cell Biol. 2018;20(12):1349–1360. doi:10.1038/s41556-018-0236-7
39. Hu W, Liu C, Bi Z, et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol Cancer. 2020;19(1):102. doi:10.1186/s12943-020-01199-1
40. Jiang X, Yang L, Gao Q, et al. The role of rab gtpases and its potential in predicting immunotherapy response and prognosis in colorectal cancer. Front Genet. 2022;13:828373. doi:10.3389/fgene.2022.828373
41. Li Y, Xiong Y, Wang Z, et al. FAM49B promotes breast cancer proliferation, metastasis, and chemoresistance by stabilizing ELAVL1 protein and regulating downstream Rab10/TLR4 pathway. Cancer Cell Int. 2021;21(1):534. doi:10.1186/s12935-021-02244-9
42. Dai S, Li N, Zhou M, et al. LncRNA EBLN3P promotes the progression of osteosarcoma through modifying the miR-224-5p/Rab10 signaling axis. Sci Rep. 2021;11(1):1992. doi:10.1038/s41598-021-81641-6
43. Ding N, Sun X, Wang T, et al. miR‑378a‑3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int J Molecul Med. 2018;42(1):381–391. doi:10.3892/ijmm.2018.3639
44. Dai J, Su Y, Zhong S, et al. Exosomes: key players in cancer and potential therapeutic strategy. Curr Signal Transduct Ther. 2020;5(1):145. doi:10.1038/s41392-020-00261-0
45. Fu W, Li T, Chen H, et al. Research progress in exosome-based nanoscale drug carriers in tumor therapies. Front Oncol. 2022;12:919279. doi:10.3389/fonc.2022.919279
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.