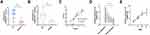Back to Journals » OncoTargets and Therapy » Volume 13
Exosomal Long Non-Coding RNA CEBPA-AS1 Inhibits Tumor Apoptosis and Functions as a Non-Invasive Biomarker for Diagnosis of Gastric Cancer
Authors Piao H, Guo S , Wang Y, Zhang J
Received 15 November 2019
Accepted for publication 25 January 2020
Published 14 February 2020 Volume 2020:13 Pages 1365—1374
DOI https://doi.org/10.2147/OTT.S238706
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Hai-yan Piao,1 Shuai Guo,2 Yue Wang,2 Jun Zhang2
1Medical Oncology Department of Gastrointestinal Cancer, Liaoning Province Cancer Hospital and Institute (Cancer Hospital of China Medical University), Shenyang City, Liaoning Province 110042, People’s Republic of China; 2Gastric Cancer Department, Liaoning Province Cancer Hospital and Institute (Cancer Hospital of China Medical University), Shenyang City, Liaoning Province 110042, People’s Republic of China
Correspondence: Jun Zhang
Gastric Cancer Department, Liaoning Province Cancer Hospital and Institute (Cancer Hospital of China Medical University), No. 44 Xiaoheyan Road, Dadong District, Shenyang City, Liaoning Province 110042, People’s Republic of China
Tel +86-18900917948
Fax +86-24-2431-5679
Email [email protected]
Aim: Traditional non-invasive diagnostic markers for gastric cancer (GC) exhibit insufficient sensitivity and specificity. Circulating exosomes are clinically useful non-invasive biomarkers for tumor diagnosis. In addition to their potential role in cancer biology, circulating long non-coding RNAs (lncRNAs) are a new class of promising cancer biomarkers. In the present study, we aimed to identify lncRNAs in circulating exosomes with potential as biomarkers for GC detection.
Methods: We compared the expression of CEBPA-AS1 between GC cells and gastric epithelial cells. The biological function of exosomal CEBPA-AS1 was determined by cell phenotype experiments and rescue assays. We also compared the expression of CEBPA-AS1 in cancerous tissue from GC patients and corresponding adjacent normal tissues, as well as the expression of CEBPA-AS1 in plasma exosomes of GC patients and healthy controls. Diagnostic accuracy was assessed by the receiver operating characteristic (ROC) curve and area under the curve (AUC).
Results: CEBPA-AS1 was highly expressed in both GC cells and in exosomes secreted by GC cells. In addition, CEBPA-AS1-containing exosomes secreted by GC cells could promote cell proliferation and inhibit apoptosis, thereby inducing the malignant behavior of GC cells. The level of CEBPA-AS1 was also significantly increased in tissues and plasma exosomes of GC patients. Stability tests showed that most plasma CEBPA-AS1 was encased in exosomes, thus avoiding degradation by RNases. We evaluated the diagnostic accuracy of exosome-derived CEBPA-AS1. The AUC value of CEBPA-AS1 in discriminating GC patients from healthy controls was 0.824, which was higher than the diagnostic accuracy of other traditional tumor biomarkers.
Conclusion: CEBPA-AS1-containing exosomes secreted from GC cells could promote cell proliferation, inhibit apoptosis, and induce GC progression, indicating that exosomal CEBPA-AS1 is involved in cell-to-cell communication in GC carcinogenesis. Exosomal CEBPA-AS1 is a promising new biomarker for clinical diagnosis of GC.
Keywords: gastric cancer, exosome, lncRNAs, CEBPA-AS1, diagnosis
Introduction
Despite its declining incidence in recent years, gastric cancer (GC) remains the fourth most common cancer type and the second leading cause of cancer-related death.1 A large number of GC patients already present with distant metastases or are at high risk of developing severe complications at the time of diagnosis. Currently, the gold standard for GC diagnosis is gastroscopy plus pathological biopsy, which is invasive and uncomfortable.2 Humoral biopsy is noninvasive, but serum tumor biomarkers used for GC diagnosis have low sensitivity and specificity.3 With the development of high-throughput sequencing technology, circulating RNA in serum or plasma presents as a novel and non-invasive diagnostic biomarker.4 However, circulating RNA is susceptible to endogenous RNases, and RNAs can also be released by circulating dysfunctional cells.
Exosomes are reported as being protective against RNA degradation, and enhance the stability of RNA in circulating blood.5 Additionally, exosomes, which are mediators of intercellular communication, can both participate in tumor formation and inhibit tumor progression. Exosomes are also an important part of the tumor microenvironment, and exosomal RNA can accurately reflect the changes occurring in cancer cells during tumor development.6 Therefore, detecting tumor-derived exosomal non-coding RNA (ncRNA) may lead to the identification of new tumor biomarkers. Several reports have highlighted the clinical significance of circulating exosomal RNAs, especially miRNAs, for GC diagnosis.7–9 However, the low quantity and specificity of circulating miRNAs greatly limits their value as diagnostic molecules, and other types of RNAs, such as long non-coding RNAs (lncRNAs), may be more suitable as diagnostic markers. However, the significance of cancer cell-derived exosomal lncRNA in plasma or serum for GC diagnosis remains unclear. LncRNAs are a class of ncRNA molecules with a length of more than 200 nucleotides with limited protein-coding capacity.10 Several studies have demonstrated that lncRNAs affect and regulate gene expression at multiple levels, including epigenetic, transcriptional, transcriptional, and post-translational levels, suggesting they may be of clinical significance in humoral GC biopsy.
In the present study, we found that GC cells can produce and release exosomes, and that lncRNA CEBPA-AS1, located at 19q13.11,11 is highly expressed in GC cell-derived exosomes, as well as in GC plasma exosomes. We further verified the potential of CEBPA-AS1 as a GC diagnostic biomarker and compared it with traditional plasma tumor markers.
Materials and Methods
Cell Lines and Cell Culture
The human normal gastric epithelial cell line GES-1 and human gastric cancer cell lines SGC-7901 and BGC-823 were obtained from China Medical University (Shenyang, China). The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL penicillin/streptomycin (Invitrogen) at 37 °C under 5% CO2 and 20% O2. All experiments were performed in triplicate with independent cell cultures.
Ethical Statement and Plasma Collection
Between January 2015 and December 2018, 281 GC patients and 80 healthy controls were enrolled in the study. All patients provided written, informed consent before surgery at the Liaoning Province Cancer Hospital & Institute. All patients underwent radical gastrectomy and D2 lymph node dissection. Patients treated with chemotherapy or radiotherapy before blood collection were excluded. Tumors were staged according to the International Union against Cancer (UICC) guidelines. The detailed clinical characteristics of the patients are provided in Table 1. The blood samples included in this study were collected into vacuum tubes with EDTA anticoagulant before surgery and pharmacotherapy and handled within 1 h after collection. Blood samples were subjected to centrifugation at 5000 rpm for 10 min at 4 °C, and the plasma was then stored at −80 °C. GC tissues and matched non-cancerous adjacent tissues were collected from 40 of the above patients.
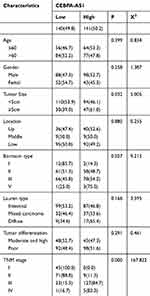 |
Table 1 CEBPA-AS1 Expressions and Clinicopathologic Characteristics |
Separation of Nuclear and Cytoplasmic Fractions
A PARIS kit (AM1921; Thermo Fisher Scientific, Yokohama, Japan) was used to divide the total cellular fractions into nuclear and cytoplasmic fractions following the manufacturer’s protocol. The kit could also be used to isolate the RNA from the same experimental sample, and was guaranteed to separate the nuclear and cytoplasmic components before the RNA was separated.
Exosome Isolation
Cells were plated in 15-cm dishes and maintained in 30 mL of RPMI-1640 medium supplemented with 10% Exosome-depleted FBS (ExoFBS™; System Biosciences, CA, USA) and 1% penicillin/streptomycin for 48 h for exosomes collection from cell culture medium. A 0.22-µm membrane filter (Millipore, USA) was used to pre-clear 20 mL of plasma (1:10 diluted in phosphate-buffered saline [PBS]) or 20 mL of culture medium. Differential centrifugation is the most common method to isolate exosomes and is considered a gold standard.12 First, the samples were centrifuged at room temperature for 20 min at 3000 × g and 10,000 × g, respectively, to remove cells and other impurities from plasma or cell culture medium. To remove microvesicles which were larger than exosomes, supernatants were then centrifuged at 100,000 × g for 30 min at 4 °C, harvested, and centrifuged again at 10,000 × g for 70 min at 4 °C. The supernatants were gently poured out and the exosome sediments were re-suspended in PBS.
Transmission Electron Microscopy
The exosome suspension was diluted to 0.5 mg/mL in PBS. The suspension was then spotted onto a glow-discharged copper grid placed on a filter paper and dried for 10 min by exposure to infrared light. Next, one drop of phosphotungstic acid (1% aqueous solution) was used to stain the exosome samples for 5 min, following which the samples were dried for 20 min by exposure to infrared light. Finally, the exosomes were visualized under a transmission electron microscope (HT7700, Hitachi, Tokyo, Japan) at 100 keV.
Western Blot Analysis
For immunoblotting, protein was extracted from an equal number of exosomes, suspended in SDS lysis buffer with proteinase inhibitors, and quantified by bicinchoninic acid assays (Pierce, USA). Then, 20 μg of exosomal protein was resolved on a 12% SDS–PAGE gel, transferred to a PVDF membrane (Millipore), and then incubated with primary antibodies against CD63 (1:1000, Abcam), CD81 (1:1000, Abcam), and HSP70 (1:1000, Santa Cruz) at 4 °C overnight. The gray values of the expression of all proteins were measured using ImageJ (NIH, Bethesda, MD, USA).
Real-Time Reverse Transcription–Polymerase Chain Reaction (Real-Time PCR)
Complementary DNA (cDNA) was generated from 500 ng of total RNA using the Promega miRNA first-strand cDNA core kit (Promega, Madison, WI, USA). Real-time PCR was performed in a LightCycler 480 (Roche AG, Basel, Switzerland) using SYBR Master Mixture (Takara Bio, Inc., Kusatsu, Japan). Each sample was analyzed in triplicate. U6 DNA was used as loading control. Fold changes in RNA expression between different cells were determined by the 2−△△CT method.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS at room temperature for 10 min and permeabilized by 0.1% Triton-X and 1% BSA in PBS for 30 min at room temperature. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Biomarkers were detected by antisense probe hybridization at 37 °C for approximately 16 h. Imaging was performed using an ECLIPSE Ni microscope (Nikon, Japan).
Lentivirus Vector System, and Cell Transfection
Lentiviruses carrying siRNA sequences targeting human CEBPA-AS1 were obtained from GeneChem (Shanghai, China). The viruses and Polybrene reagent (Abbott Laboratories, Chicago, IL, USA) were used to infect cells. GC cells were cultured for 72 h in medium containing puromycin for cell screening. For in vitro exosome assays, 1 µg of exosomes (equivalent to those collected from ~5 × 106 producer cells) were added to 2 × 105 recipient cells.
Gene Expression in TCGA
The online Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn)13 web tool was used to analyze RNA sequencing expression data which was based on The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects.
Proliferation Assay Using Cell Counting Kit-8
Cell Counting Kit-8 (CCK-8) was used to evaluate the cellular proliferative capacity according to the manufacturer’s instructions (Dojindo Laboratories, Kumamoto, Japan). The optical density at 450 nm was determined on a microplate reader (Bio-Rad, Hercules, CA, USA).
Cell Cycle and Apoptosis Analyses
Cells were cultured in 6-cm plates for 72 h at 1 × 106 cells/dish. Cells were collected for cell cycle analysis and precooled with 70% ethanol at 4 °C overnight. The cells were then washed with precooled PBS and incubated with propidium iodide (Sigma–Aldrich, St. Louis, MO, USA) at 37 °C for 1 h. For apoptosis analysis, cells were incubated with Annexin V–APC (Yeasen Biotech, Shanghai, China) at room temperature in the dark for 30 min. BD flow cytometry (Franklin Lake, New Jersey, USA) was used in both tests and FlowJo software (Ashland) was used for analysis.
Statistical Analysis
Statistical Package for Social Science (SPSS; IBM, Armonk, NY, USA) version 19.0 was used for statistical analysis. All data are presented as means ± SD (standard deviation). The Student’s t-test was used when the variance between groups was similar; otherwise, the Wilcoxon-signed rank test was used. One-way analysis of variance (ANOVA) was used for comparisons between multiple groups. Differences with a P-value <0.05 were considered significant.
Results
The Expression of CEBPA-AS1 in GC Cells and Exosomes
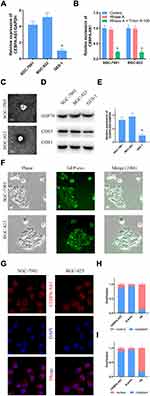
qPCR results showed that the expression of CEBPA-AS1 was increased in BGC-823 and SGC-7901 cells compared with that in control GES-1 cells (Figure 1A). Next, we investigated whether extracellular CEBPA-AS1 could be detected. The level of CEBPA-AS1 in culture medium (CM) did not change with RNase treatment alone; however, the level of CEBPA-AS1 significantly decreased when the CM was treated with RNase plus Triton x-100 (Figure 1B). This indicated that extracellular CEBPA-AS1 was mainly enveloped by membrane, rather than being directly released. Exosomes isolated from GC cells had a classical cup shape, size, and number (Figure 1C). Although both GC cells and control cells could secrete exosomes (Figure 1D), the level of exosomal CEBPA-AS1 derived from GC cells was significantly higher than that of control cells (Figure 1E). GC cells were co-incubated with GC cell-derived exosomes which were labeled fluorescent green. Fluorescence microscopy analysis showed that the exosomes could be taken up by GC cells (Figure 1F).
Fluorescence microscopy analysis further showed that the CEBPA-AS1 signal was mainly localized in the cytoplasm of GC cells (Figure 1G). Moreover, a similar result was obtained with nuclear/cytoplasmic RNA fractionation (Figure 1H and I).
Exosomal CEBPA-AS1 Expression Affects the Malignant Phenotype of GC Cells
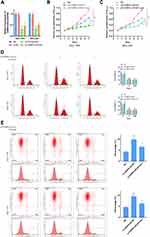
Based on the above analysis, we reasoned that the abnormal expression of CEBPA-AS1 could be closely related to the malignant phenotype of GC cells. Therefore, we evaluated the biological role of CEBPA-AS1 in vitro. As CEBPA-AS1 was highly expressed in GC cells, we used RNAi to silence the expression of CEBPA-AS1 (Figure 2A). Si-CEBPA-AS1#1 inhibited the expression of CEBPA-AS1 to the greatest extent and was therefore selected for subsequent studies. We also isolated exosomes from untreated SGC-7901 and BGC-823 cells, which we called CEBPA-AS1-exo. We added si-CEBPA-AS1#1 and/or exosome-riched CEBPA-AS1 at 40 μg/mL to GC cells, and investigated the effect of CEBPA-AS1 on cell proliferation using CCK-8 assays. The results showed that CEBPA-AS1 knockdown significantly inhibited the proliferative capacity of GC cells, while this effect could be partially reversed in both types of GC cells when CEBPA-AS1-exo was added to the medium (Figure 2B and C). Additionally, si-CEBPA-AS1#1 induced cycle arrest of GC cells at the S and G2 phases, but this effect was also partially rescued by the addition of CEBPA-AS1-exo, which induced an increase in the percentage of cells in the G1 phase (Figure 2D). Similarly, si-CEBPA-AS1#1 increased the number of apoptotic GC cells, which again was reversed by co-exposure to CEBPA-AS1-exo (Figure 2E). These results indicated that CEBPA-AS1 contributed to the malignant phenotype of GC cells and exosomal CEBPA-AS1 served as a mediator in intercellular communication.
We also downregulated CEBPA-AS1 expression in GES-1 cells by RNAi, and found that apoptosis (Supplementary Figure 1A and B), cell cycle (Supplementary Figure 1C and D), and proliferation (Supplementary Figure 1E) were all affected, although not significantly. Again, this effect was rescued by CEBPA-AS1-exo application.
The flank 10 k theory suggests that lncRNAs can cis-regulate adjacent genes within a range of 10,000 bp of its chromosome location.14 Therefore, we investigated whether CEBPA-AS1 could affect the expression of CEBPA (CCAAT enhancer binding protein alpha) using GEPIA, an interactive web server based on TCGA. The TCGA expression profile data showed that CEBPA was highly expressed in GC patients (Supplementary Figure 2A). In addition, qPCR data indicated that CEBPA was also highly expressed in GC cells (Supplementary Figure 2B). Moreover, once the cells had absorbed the CEBPA-AS1-exo, the expression of CEBPA continued to rise (Supplementary Figure 2C). This suggested that the effect of CEBPA-AS1 on the malignant phenotype of GC was exerted through CEBPA, similar to that reported by Guo Y et al11 for oral squamous cell carcinoma.
CEBPA-AS1 Is Highly Expressed in GC Tissues and GC-Derived Exosomes
We then measured the expression of CEBPA-AS1 in tissue samples and that of CEBPA-AS1-exo in plasma samples from patients. Forty matched specimens were included in the evaluation. CEBPA-AS1 was expressed in all GC tissue samples, and its expression level was higher in GC samples than in adjacent non-cancerous tissues (Figure 3A). Then, we enrolled 281 GC patients and 80 normal healthy subjects. The levels of exosomal CEBPA-AS1 in plasma samples from the 281 patients were measured by qPCR. Exosomal CEBPA-AS1 expression levels were markedly upregulated in plasma samples derived from GC patients compared with those from healthy subjects (Figure 3B, P < 0.01).
Forty GC patients’ tissue and plasma sample were used to determined whether a correlation existed between CEBPA-AS1 in tissue and exosomal CEBPA-AS1 in plasma. It found that a significant positive correlation between the two groups (Figure 3C, R = 0.6756, P < 0.01). Then, we measured the levels of CEBPA-AS1 in plasma exosomes derived from the 281 GC patients before and after surgery. The results showed that the level of plasma exosomal CEBPA-AS1 decreased significantly after surgery (Figure 3D, P < 0.01). We also found that CEBPA-AS1 expression in plasma exosomes was related to TNM stage, and that the expression of CEBPA-AS1 increased with increasing tumor malignancy (Figure 3E, P < 0.05).
Analysis of CEBPA-AS1 Stability in Plasma
The stability of circulating RNA is a prerequisite for its diagnostic capacity. To test the stability of CEBPA-AS1 in plasma, 10 GC samples were directly treated with RNase or RNase plus Triton X-100. Similar to the in vitro results, CEBPA-AS1 levels in plasma did not change after RNase treatment alone (Figure 4A). However, the expression of CEBPA-AS1 was significantly reduced with combined RNase and Triton x-100 treatment and the Ct value was significantly increased (Figure 4B). Finally, the plasma samples were stored at −80 °C for 3 months with no significant change in Ct value (Figure 4C). This suggested that the expression of CEBPA-AS1 in plasma exosomes was stable.
Correlation Between the Expression of Exosomal CEBPA-AS1 and Clinicopathological Features
The patients were divided into high-expression and low-expression groups based on the median expression levels of CEBPA-AS1 in the 281 enrolled patients. The relationship between clinicopathological features and CEBPA-AS1 expression is shown in Table 1. Chi-square tests showed that the expression of CEBPA-AS1 was closely related to tumor size (P = 0.032, χ2 = 5.005), Bormann type (P = 0.027, χ2 = 9.215), and TNM stage (P = 0.000, χ2 = 167.822).
Diagnostic Value of Exosomal CEBPA-AS1 for GC Patients
CEA, CA19-9, CA72-4, CA12-5, and AFP are widely used for screening and auxiliary diagnosis of malignant tumors of the digestive system. The ROC curve and AUC value of plasma exosomal CEBPA-AS1 was significantly higher than that of these traditional markers (Figure 5A–F). The combined diagnostic AUC value of these markers (Figure 5G) was higher than that of each single marker, but still lower than that of plasma exosome CEBPA-AS1. Finally, we evaluated the diagnostic value of CEA, CA19-9, CA72-4, CA12-5, AFP, and exosome CEBPA-AS1 for GC, and found that the AUC value was higher than any single biomarker (Figure 5H). When 2.514 was selected as the best cut-off value, the sensitivity and specificity of exosomal CEBPA-AS1 were 87.9% and 78.8%, respectively. Overall, these data suggest that circulating exosomal CEBPA-AS1 is a potential biomarker for GC diagnosis.
Discussion
Gastric cancer is a common digestive system malignant tumor with complex etiology. Because of the low rate of early diagnosis, most GC patients are diagnosed at an advanced stage in China. Although traditional treatment strategies have improved, the 5-year survival rate is still low.15 Therefore, it is critical that GC is diagnosed early, and finding a suitable plasma molecular biomarker may be an effective method to achieve this goal.
Several studies have demonstrated the feasibility of using circulating lncRNAs as biomarkers for tumor diagnosis; however, numerous preanalytical and analytical aspects, such as sample choice, extraction method, and donor-related factors, may affect the accuracy of circulating lncRNA determination. The stability and longevity of lncRNA molecules are preconditions for cancer noninvasive diagnosis.16 Exosomes contain various cargos, and are ideal carriers owing to their double-membrane structure. Exosomes secreted by tumor cells can deliver pathological substances to normal cells, thereby affecting the growth and differentiation of normal cells. In addition, some exosomes can enter the blood circulation as potential molecular diagnostic markers or participate in tumor invasion and metastasis.17
CEBPA-AS1, also known as LOC80054, was first detected in a sample of 51 GC patients. Together with the other four lncRNAs, they exhibit diagnostic value in GC.18 Furthermore, CEBPA-AS1 was also highly expressed in oral squamous cell carcinoma (OSCC) tissue, and may participate in the development of OSCC through the CEBPA-AS1/CEBPA/BCL2 axis.11 TCGA data mining revealed that CEBPA-AS1 participated in the establishment of a lung adenocarcinoma diagnostic prediction model.19 Although CEBPA-AS1 may be an oncogene, its presence in tumor-derived exosomes has not been reported. Our results showed that CEBPA-AS1 was enriched in GC cells, and could be secreted by exosomes. Moreover, exosomes containing CEBPA-AS1 affected the proliferation and apoptosis of GC cells. This implies that CEBPA-AS1 is related to the malignant biological behavior of GC, and has diagnostic significance. Stable expression is the premise for the clinical application of exosomal CEBPA-AS1 as a diagnostic marker. We found that plasma CEBPA-AS1 could not be degraded by RNase, and CEBPA-AS1 could be stable for extended periods when stored at −80 °C. The diagnostic efficacy of plasma exosomal CEBPA-AS1 was significantly higher than that of conventional tumor biomarkers, and was closely related to GC clinical stage.
The study of exosomal lncRNAs is still in its infancy. At present, many standards have not been unified, such as whole blood collection, preparation, treatment, and storage of plasma/serum, exosome and RNA extraction, lncRNA quantification, and the cut-off value of lncRNA. Only consensus in these areas can reduce the differences in results between laboratories that may have a significant impact on lncRNA results. Despite the above difficulties, exosomal lncRNAs have great potential as biomarkers. First, exosomes exhibit specificity, and possess unique markers for easy identification, and the contents released by exosomes can differ under different physiological or pathological conditions.20 LncRNAs have a selective loading mechanism, namely, lncRNA expression is lower in cells than in exosomes.21 Secondly, lncRNA stability is not affected by external conditions as it is protected by a lipid bilayer.22 Therefore, investigation into exosomal lncRNAs deserves our time and energy.
Although we identified and verified the importance of CEBPA-AS1 exosomes in the diagnostic significance of GC, the present study nevertheless has some limitations. First, we did not identify the mechanism associated with the role of CEBPA-AS1 in GC pathogenesis. Second, patients who received neoadjuvant chemotherapy were not included in this study, and the diagnostic significance of CEBPA-AS1 for this group of patients remains unclear, including whether CEBPA-AS1 expression is decreased with disease remission. Third, we did not analyze the relationship between CEBPA-AS1 expression and survival in GC. These questions need to be addressed in future studies.
Conclusion
In conclusion, our study demonstrated the presence of CEBPA-AS1 in exosomes isolated from GC cells culture medium and plasma samples of GC patients, and it was associated with GC proliferation and apoptosis. In addition, it was overexpressed in GC culture medium and plasma. Compared with traditional tumor markers, CEBPA-AS1 may be a better diagnostic marker.
Ethics and Informed Consent
The study was reviewed and approved by the Faculty of Science Ethics Committee at Liaoning Cancer Hospital & Institute (Cancer Hospital of China Medical University)(20181226). All patients provided written informed consent before surgery at the Liaoning Province Cancer Hospital and Institute and that this was conducted in accordance with the Declaration of Helsinki.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Author Contributions
HYP performed the majority of experiments and analyzed the data and drafted the manuscript; JZ designed the research; SG conducted the molecular biology assays and assisted in writing the manuscript; YW collected and analyzed the data. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Park JY, von Karsa L, Herrero R. Prevention strategies for gastric cancer: a global perspective. Clin Endosc. 2014;47(6):478–489. doi:10.5946/ce.2014.47.6.478
3. Acharya A, Markar SR, Matar M, et al. Use of tumor markers in gastrointestinal cancers: surgeon perceptions and cost-benefit trade-off analysis. Ann Surg Oncol. 2017;24(5):1165–1173. doi:10.1245/s10434-016-5717-y
4. Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39. doi:10.1186/s12943-016-0524-4
5. Xu H, Chen Y, Dong X, et al. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27(6):710–716. doi:10.1158/1055-9965.epi-17-0770
6. Fan Q, Yang L, Zhang X, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi:10.1016/j.canlet.2017.10.040
7. Kumata Y, Iinuma H, Suzuki Y, et al. Exosomeencapsulated microRNA23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol Rep. 2018;40(1):319–330. doi:10.3892/or.2018.6418
8. Huang Z, Zhu D, Wu L, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(2):188–196. doi:10.1158/1055-9965.epi-16-0607
9. Shekari N, Baradaran B, Shanehbandi D, et al. Circulating MicroRNAs: valuable biomarkers for the diagnosis and prognosis of gastric cancer. Curr Med Chem. 2018;25(6):698–714. doi:10.2174/0929867324666171003123425
10. Lau E. Non-coding RNA: zooming in on lncRNA functions. Nat Rev Genet. 2014;15(9):574–575. doi:10.1038/nrg3795
11. Guo Y, Ma Y, Hu X, et al. Long non-coding RNA CEBPA-AS1 correlates with poor prognosis and promotes tumorigenesis via CEBPA/Bcl2 in oral squamous cell carcinoma. Cancer Biol Ther. 2018;19(3):205–213. doi:10.1080/15384047.2017.1416276
12. Momen-Heravi F, Balaj L, Alian S, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394(10):1253–1262. doi:10.1515/hsz-2013-0141
13. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
14. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
15. Zhang J, Wu Y, Lin YH, et al. Prognostic value of hypoxia-inducible factor-1 alpha and prolyl 4-hydroxylase beta polypeptide overexpression in gastric cancer. World J Gastroenterol. 2018;24(22):2381–2391. doi:10.3748/wjg.v24.i22.2381
16. Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–85563. doi:10.18632/oncotarget.13465
17. Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28(5):1551–1558. doi:10.3892/or.2012.1967
18. Ke D, Li H, Zhang Y, et al. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8(13):21516–21525. doi:10.18632/oncotarget.15628
19. Sui J, Yang S, Liu T, et al. Molecular characterization of lung adenocarcinoma: a potential four-long noncoding RNA prognostic signature. J Cell Biochem. 2019;120(1):705–714. doi:10.1002/jcb.27428
20. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27(3):172–188. doi:10.1016/j.tcb.2016.11.003
21. Gezer U, Ozgur E, Cetinkaya M, et al. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi:10.1002/cbin.10301
22. Yang Y, Cai Y, Wu G, et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci (Lond). 2015;129(8):675–685. doi:10.1042/cs20150121
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.