Back to Journals » Journal of Inflammation Research » Volume 14
Exosomal lncRNA HOTTIP Mediates Antiviral Effect of Tenofovir Alafenamide (TAF) on HBV Infection
Authors Liu QM, He YY, Liu LL, Wang LK
Received 14 April 2021
Accepted for publication 19 September 2021
Published 23 October 2021 Volume 2021:14 Pages 5489—5500
DOI https://doi.org/10.2147/JIR.S315716
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Monika Sharma
Qing-Min Liu,1,* Yi-Yu He,2,* Li-Li Liu,3 Li-Kun Wang4
1Intensive Care Unit, Linyi People’s Hospital, Linyi, Shandong Province, People’s Republic of China; 2Department of Cardiovascular Disease, Renmin Hospital of Wuhan University, Wuhan, Hubei Province, People’s Republic of China; 3Department of Pathology, Linyi People’s Hospital, Linyi, Shandong Province, People’s Republic of China; 4Infection Control Center, Linyi People’s Hospital, Linyi, Shandong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li-Li Liu
Department of Pathology, Linyi People’s Hospital, Linyi, Shandong Province, People’s Republic of China
Email [email protected]
Li-Kun Wang
Infection Control Center, Linyi People’s Hospital, Linyi, Shandong Province, People’s Republic of China
Email [email protected]
Introduction: Chronic hepatitis B (CHB) virus (HBV) infection has emerged as a global health burden affecting nearly 292 million people. Tenofovir alafenamide (TAF) is an effective treatment for CHB patients. However, the detailed mechanism underlying the antiviral activity of TAF remains unclear.
Methods: In this study, we investigated the antiviral effect of exosomes derived from the serum of CHB patients treated with TAF (Exo-serum) and TAF-treated macrophages (MP) (Exo-MP(TAF)).
Results: RNAseq analysis was also performed to determine the associated long non-coding RNAs (lncRNAs). The results demonstrated that both Exo-serum and Exo-MP(TAF) could be taken up by HepAD38 cells and exhibited potent antiviral activities, as manifested by significantly downregulating the levels of hepatitis B surface antigen, hepatitis B e antigen, HBV DNA, and covalently closed circular DNA. The antiviral effect of Exo-serum was more potent than those of TAF treatment alone. RNAseq analysis revealed that lncRNA HOTTIP was upregulated significantly in Exo-serum. Further, lncRNA HOTTIP knockdown reversed the antiviral effect of Exo-MP(TAF) on HepAD38 cells, whereas lncRNA HOTTIP knockdown exerted the opposite roles.
Discussion: Taken together, these results suggest that exosomal lncRNA HOTTIP is essential for the antiviral activity of TAF and provide a novel understanding of the exosome-mediated mechanism underlying HBV infection.
Keywords: chronic hepatitis B, tenofovir alafenamide, exosome, lncRNA HOTTIP
Introduction
Chronic hepatitis B (CHB) virus (HBV) infection is a leading cause of chronic liver diseases, such as liver cirrhosis and hepatocellular carcinoma (HCC).1 CHB has emerged as a global health burden, impacting nearly 292 million people.2 In China, around 86 million people are HBV carriers, accounting for 25% of the global total, and approximately 32 million patients suffer from CHB.3 At present, there are two major types of treatments, nucleos(t)ide analogues (NAs) and interferons (IFNs), which are effective in inhibiting HBV replication and reducing the risk of developing into more severe liver diseases.4 Among the NAs, tenofovir alafenamide (TAF) is a phosphonamidate oral prodrug of tenofovir for the treatment of CHB.5 TAF is stable in the plasma and exerts a potent antiviral effect against HBV with a high barrier to antiviral resistance, allowing TAF to be a preferred option for treating patients with CHB.6,7 However, the mechanism underlying the antiviral effect of TAF remains to be elucidated in detail.
Exosomes, extracellular vesicles 30 to 150 nm in diameter, originate from endosomal multivesicular bodies.8 It has been demonstrated that exosomes are critical mediators in intercellular communication for transporting functional molecules, including proteins, DNAs, and non-coding RNAs (ncRNAs), from donor cells to recipient cells.9 Thus, the essential function of exosomes has been extensively observed in various biological processes, such as cancer initiation and progression, cellular homeostasis, and immune response.10 Regarding HBV infection, exosomes exert an important role in HBV transmission and immune regulation of HBV infection.11 Furthermore, exosomal microRNA (miRNA)-122 and miRNA-146a are upregulated and downregulated in the serum of CHB patients, respectively.12 In another study, exosomal miRNA-574-5p derived from interferon-treated macrophage (MP) is associated with antiviral activity for HBV infection.13 It is suggested that exosomal molecules are potential regulators in regulating HBV infection.
Long non-coding RNAs (lncRNAs), a class of ncRNAs, are over 200 nucleotides in length without protein-coding capability.14 So far, numerous studies have reported that lncRNAs are vital regulators in biological processes, such as cell proliferation, signaling transmission, and apoptosis.15 In addition, the role of exosomal lncRNAs has also been uncovered in various intercellular communication mechanisms.16 To the best of our knowledge, however, the effect of exosomal lncRNA on HBV infection has not been investigated. Thus, this study aimed to elucidate the effect of exosomes on TAF-induced antiviral effect against HBV infection and the underlying mechanisms.
Materials and Methods
Ethics Statement
The study was performed according to Good Clinical Practice and the ethical principles of the Declaration of Helsinki17 and the STROBE statement. Experimental protocols and ethics approval forms were approved by the Ethical Review Board of Linyi People’s Hospital (BS201479-C). Written, informed consents were obtained from the parents of patients with CHB.
Samples
Six patients (average age 38.4 years) were diagnosed with CHB infection. All enrolled patients had treatment switching from NAs to TAF monotherapy and had at least a 1-year course of entecavir (ETV) or an NA combination of lamivudine (LAM)/tenofovir disoproxil fumarate (TDF), ETV/adefovir (ADF), or ETV/TDF. Patients took a dose of 25 mg TAF orally daily (Vemlidy; Gilead Sciences KK, Tokyo, Japan). Exclusion criteria were as follows: (a) hepatocellular carcinoma; (b) decompensated cirrhosis; (c) terminal illness; (d) positivity for antibody to HIV, hepatitis C, and hepatitis D; (e) without sufficient medical records; (f) other severe chronic diseases. Serum samples were collected from these patients at 0, 24, and 48 weeks post-TAF treatment. The clinical characteristics of the CHB patients are listed in Table 1.
 |
Table 1 The Clinical Characteristics of the CHB Patients |
Measurement of HBsAg, Hepatitis B e Antigen, HBV DNA, and HBV cccDNA
Hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) were quantified using electrochemiluminescence on a cobas e 601 instrument (Roche Diagnostics, USA). HBV-DNA level was quantified using fluorescence RT-PCR on the ABI7500 platform (Applied Biosystems, USA). Intracellular HBV covalently closed circular DNA (cccDNA) was quantified using Light Cycler Fast Start DNA Master HybProbe (Roche Diagnostics, USA) according to the manufacturer’s instructions. Meanwhile, the level of cccDNA was also determined by the Southern blot assay as previously described.18
Exosome Extraction
Exosomes isolation was performed as previously described.19 Briefly, serum samples (1 mL) were centrifuged at 300 ×g for 10 min and were diluted with sterile PBS solution in 1:1 ratio. Then, the samples were centrifuged at 10,000 ×g for 30 min, followed by ultracentrifugation at 200,000 ×g for 2 h. The exosome pellets were washed with PBS solution three times and filtered through syringe filters (0.2 μm). Afterward, the samples were centrifuged at 200,000 ×g for 1 h and resuspended on PBS. All centrifugation processes were carried out at 4°C. Exosomes were quantified using Pierce BCA Protein Assay Kit according to the manufacturer’s instructions (Thermo Fisher Scientific, USA).
Transmission Electron Microscopy (TEM)
Exosomes were resuspended in PBS solution and placed on copper carbon-supported grids (Electron Microscopy Sciences, USA). After 5-min incubation at room temperature, exosomes were fixed with 2% paraformaldehyde. The grids were stained with 10% uranyl acetate for 10 min at room temperature. Exosomes were photographed using a JEOL 100XCII electron microscope (JEOL, USA).
Nanoparticle Tracking Analysis (NTA)
Nanoparticle tracking analysis was used to determine the concentration and size distribution. NTA was performed on Nanosight NS300 system (Malvern Instruments Company, UK) as previously described.20
Western Blotting
Exosomes (5 µg) were lysed with cell lysis buffer (Beyotime Institute of Biotechnology, China). Protein extracts were separated by SDS-PAGE electrophoresis and were transferred electrically to Polyvinylidene difluoride (PVDF) membranes (Thermo Fisher Scientific, USA). The samples were blocked with 5% skim milk in PBS-0.05% Tween 20 for 2 h at room temperature. Afterward, the primary antibodies against GAPDH (1;1000), CD81 (1;1000), and TSG101 (1:1000) (Abcam, USA) were applied overnight at 4°C. Then, the membranes were incubated with their respective secondary antibody for 1 h at 37°C and visualized by enhanced chemiluminescence assays (Thermo Fisher Scientific, USA). The quantification of optical density for bands was determined via Imagej software (version 1.8).
Cell Cultures
HepAD38 cells with stable HBV replication were kindly provided by Peking University. THP-1 cells were obtained from American Type Culture Collection (Manassas, USA). HepAD38 cells were maintained in Dulbecco’s modified Eagle medium (Gibco, USA) supplemented with 100 μg/mL streptomycin, 100 U/mL penicillin, and 10% fetal bovine serum. THP-1–derived MP were cultured in Roswell Park Memorial Institute 1640 medium (Gibco, USA) supplemented with 100 μg/mL streptomycin, 100 U/mL penicillin, and 10% extracellular vesicle-free fetal bovine serum. 50 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich, USA) was applied to induce cell differentiation for 24 h. All cells were cultured at 37°C and 5% CO2. TAF treatment with indicated concentrations was applied to HepAD38 cells for 48 h, and then cells were collected for subsequent experiments.
Fluorescence Staining Assay
Exosomes were stained with PKH67 Green Fluorescent Cell Linker Kit according to the manufacturer’s instructions (Sigma-Aldrich; USA) and were cocultured with HepAD38 cells for 8 h. Then, the coculture system was labeled with fluorescent dyes rhodamine phalloidin and Hoechst (Thermo Fisher Scientific, USA). The images were taken using a confocal microscope (Carl Zeiss LSM710, Germany)
RNAseq
Total RNAs were isolated from serum samples using exoRNeasy Serum/Plasma Maxi Kit according to the manufacturer’s instructions (Qiangen, Germany). RNA quantity was determined using NanoDrop™ 2000/2000c Spectrophotometers (Thermo Fisher Scientific, USA). RNAseq assays, including RNA quality control, library preparation, and sequencing, were carried out by Nanodigmbio Biotechnology Co. LTD (Shanghai, China). RAN sequencing was performed on Illumina HiSeq2500 system (Illumina, USA). Raw 125 bp reads were obtained, and low-quality reads were removed before analysis. Bioinformatics analyses were performed as previously described.21 The raw data were stored in the Sequence Read Archive (SRA) (SUB9894356).
RT-PCR
Total RNA was isolated from exosomes or cells using the RNeasy Mini kit (Qiagen, Germany) following the manufacturer’s instructions. RNA quantify was determined using NanoDrop™ 2000/2000c Spectrophotometers (Thermo Fisher Scientific, USA). Then, the first-strand cDNA was synthesized using a reverse transcription PCR kit (Applied Biosystems, USA) according to the manufacturer’s instructions. RT-PCR was performed on the ABI7500 platform (Applied Biosystems, USA) according to the required conditions for each primer. Raw data were using the 2−ΔΔCt method22 with β-actin as the reference gene.
Transfections
Small interfering RNA (siRNA) against lncRNA HOTTIP (siRNA-HOTTIP) and negative control (siRNA-NC) were purchased from Shanghai GenePharma Co, Ltd. The sequences were as follows: siRNA-HOTTIP: 5′-AAAUUGCUCACUAACAGUGUG-3′; siRNA-NC: 5′-GGUGGAACAAUUGCUUUUA-3′. The plasmids carrying the coding sequence of lncRNA HOTTIP were constructed using KOD-plus-Ver.2 kit (Toyobo Life Science, Japan) and then cloned into pcDNA3.1 vector according to the manufacturer’s instructions. When cells reached about 80% confluent in 6-well plate, cell transfection assay was conducted using Lipofectamine® 3000 (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. 48 h post-transfection, cells were collected for subsequent experiments.
CCK-8 Assay
Cells (1 × 104 cells/well) were seeded on 96-well plates and were subjected to the indicated treatment. Cell viability was determined via CCK-8 assays (Dojindo Laboratories, Japan) according to the manufacturer’s instruction. The raw data were obtained using a microtiter plate reader at 450 nm absorbance. The results were the mean of three independent triplicates.
Statistical Analysis
Data were presented as mean ± SD. Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software, USA). Differences between groups were analyzed with Student’s t-test. Differences were considered to be significant at p < 0.05, denoting as * p < 0.05, ** p < 0.01, *** p < 0.001.
Results
Characterization of Exosomes Derived from the Serum of CHB Patients
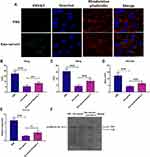
In this study, 6 CHB patients who had already received daily TAF treatment (25 mg) for 48 weeks were enrolled. The data showed that TAF treatment dramatically reduced the serum HBsAg levels at 24 and 48 weeks after receiving TAF treatment, compared with those at day 0 (Figure 1A). Then, extracellular vesicles (EVs) were isolated from the serum of these 6 CHB patients. The Western blotting assay illustrated that EVs positively expressed exosomal markers CD81 and TSG101 (Figure 1B). Using TEM and NTA assays, the results further revealed that EVs displayed classical bilayer round shape, with 139 nm in diameter (Figure 1C). Collectively, these data demonstrated that EVs isolated from the serum of CHB patients were exosomes (Exo-serum).
The Antiviral Effect of Exosomes Derived from the Serum of CHB Patients
Next, we applied green fluorescent dye PKH67 to label Exo-serum and found that Exo-serum could be taken up by HepAD38 cells under confocal fluorescence microscopy (Figure 2A). Furthermore, after coculturing with Exo-serum for 48 h, the levels of HBsAg, HBeAg, and HBV DNA were downregulated in the culture supernatants of HepAD38 cells, relative to those treated with PBS (Figure 2B–D). A reduction of intracellular HBV cccDNA level of HepAD38 cells was also observed after treating with Exo-serum (Figure 2E and F). Meanwhile, the antiviral effect of Exo-serum was partially reversed by the application of Annexin V, a suppressor for exosome internalization.23 Together, Exo-serum could be internalized and play an antiviral role in HepAD38 cells.
Comparison of Antiviral Effects of Exosomes Derived from the Serum of CHB Patients with TAF Treatment
To investigate the effect of TAF treatment, we first applied different concentrations of TAF to treat HepAD38 cells, ranging from 0.5 to 500 μmol/L. The results suggested that TAF at concentrations of 0.5 and 5 μmol/L did not cause a significant antiviral effect (Figure 3A–D). On the other hand, the concentration of 50 μmol/L or beyond displayed antiviral activity in HepAD38 cells, as manifested by decreasing HBsAg, HBeAg, and HBV DNA levels in the culture supernatants of HepAD38 cells and inhibiting intracellular HBV cccDNA level (Figure 3A–D). Meanwhile, the concentration of 50 μmol/L and lower ones did not impact the cell viability, whereas 250 μmol/L of TAF profoundly inhibited cell viability, as illustrated by the CCK-8 assay (Figure 3E). Given these observations, the lowest effective concentration (50 μmol/L) of TAF was used in subsequent experiments. Afterward, we employed Exo-serum (10 μg/mL) and TAF treatment (50 µmol/L) to treat HepAD38 cells for 48 h. The results demonstrated that TAF treatment profoundly suppressed the levels of HBsAg, HBeAg, HBV DNA, and intracellular HBV cccDNA. Meanwhile, a more potent antiviral effect was observed in HepAD38 cells treated with Exo-serum (Figure 3F–H).
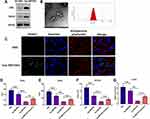
Characterization of Exosomes Derived from Macrophages Treated with TAF
In the next experiment, we applied TAF treatment (50 µmol/L) to treat THP-1-derived MP and isolated EVs derived from the culture supernatants of MP. The Western blotting assay revealed that EVs are highly expressed exosomal markers CD81 and TSG101 (Figure 4A). Also, the round morphology of EVs, with 141 nm in diameter, was confirmed by TEM and NTA assays (Figure 4B). Thus, these data indicated that EVs derived from the culture supernatants of MP treated with TAF were exosomes (Exo-MP(TAF)). Using the fluorescence staining assay mentioned above, we found that Exo-MP(TAF) could be internalized by HepAD38 cells under confocal fluorescence microscopy (Figure 4C). Furthermore, we observed that Exo-MP(TAF) played a more potent antiviral role in HepAD38 cells, compared with exosomes derived from the supernatants of THP-1-derived MP treated with PBS (Exo-MP(Exo-MP (PBS)) and PBS (Figure 4D–G). Meanwhile, the addition of Annexin V could partially attenuate the antiviral effect of Exo-MP(Exo-MP (TAF) on HepAD38 cells (Figure 4D–G). Collectively, these data suggest that exosomes may play an essential role in the antiviral effects of TAF treatment.
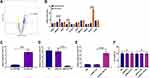
LncRNA HOTTIP Was Significantly Upregulated in Exosomes Derived from the Serum of CHB
LncRNAs have been demonstrated to be critical regulators for HBV infection24 and are vital molecules transferred by exosomes in intercellular communication.16 To investigate the potential lncRNAs associated with the antiviral effect of Exo-serum, we applied RNAseq analysis to screen the differentially expressed lncRNAs between Exo-serum and exosomes derived from the serum of CHB patients before treating with TAF treatment (Exo-serum-NC). As shown in Figure 5A, lncRNA HOTTIP was one of the most upregulated lncRNAs in Exo-serum. Using the RT-PCR assay, we measured the top 10 upregulated lncRNAs obtained from RNAseq analysis and found that only three lncRNAs, including HOTTIP, KRASP1, and H19, were increased in Exo-serum compared with Exo-serum-NC (Figure 5B). Additionally, we observed that lncRNA HOTTIP displayed the highest upregulation (Figure 5C). Thus, we focused on the effect of lncRNA HOTTIP on the antiviral effect of Exo-serum. Next, we determined the expression of lncRNA HOTTIP in Exo-MP(TAF) and Exo-MP(PBS) and found that lncRNA HOTTIP was upregulated in Exo-MP(TAF) relative to Exo-MP(PBS). This indicated that lncRNA HOTTIP might also be a critical factor for the antiviral role of TAF treatment. Then, we aimed to investigate the role of lncRNA HOTTIP further. We employed siRNA against lncRNA HOTTIP (siRNA-HOTTIP) and pcDNA3.1 vector carrying lncRNA HOTTIP (pcDNA3.1-HOTTIP) to knockdown and overexpress lncRNA HOTTIP successfully, respectively (Figure 5D and E). Also, the CCK-8 assay demonstrated that the transfection of siRNA-HOTTIP and pcDNA3.1-HOTTIP did not alter cell viability (Figure 5F).
LncRNA HOTTIP Was Essential for the Antiviral Effect of Exosomes Derived from Macrophages
In a series of experiments, HepAD38 cells were firstly treated with Exo-MP(Exo-MP (TAF) for 48 h and then treated with siRNA-HOTTIP or siRNA negative control (siRNA-NC). The results indicated that the lncRNA HOTTIP knockdown partially reversed the effect of Exo-MP(Exo-MP (TAF) on HepAD38 cells (Figure 6A–H). On the contrary, the overexpression of lncRNA HOTTIP further enhanced the antiviral effect of Exo-MP(Exo-MP (TAF). As such, these observations demonstrated that lncRNA HOTTIP is a crucial regulator mediating the antiviral effect of Exo-MP(Exo-MP (TAF).
Discussion
TAF is a newly approved prodrug of tenofovir with higher levels of the effective metabolite tenofovir-diphosphate and lower level of plasma tenofovir.25 These advantages allow TAF to be administered at a much lower dose than other treatments, for example, TDF, further lowing systemic tenofovir exposures.26 In several clinical trials, TAF displays enhanced safety profiles and equivalent efficacy in bone and renal toxicity in HBV-infected patients,7,27,28 indicating TAF is more suitable for patients with underlying renal or bone disease. Moreover, it has been well documented that TAF displays a potent antiviral effect against HBV infection, as manifested by HBV DNA suppression, HBsAg clearance, and HBeAg loss.29,30 As the significant barrier for treating HBV, the persistent cccDNA can lead to a viral relapse after antiviral treatment.31 Intriguingly, our study reported that Exo-serum could exhibit potent antiviral activity against HBV infection through inhibiting levels of HBsAg, HBeAg, HBV DNA, as well as cccDNA. Also, our report demonstrated that the antiviral effect of Exo-serum was more potent than those of TAF alone. These results together suggest that the exosome-involved mechanism is essential for the antiviral activity of TAF, and exosome-based treatments might be a promising therapeutic strategy for CHB patients.
So far, it has been well illustrated that exosomes participate in regulating HBV infection, such as intracellular HBV replication and immune response coping with HBV infection.32 For example, several enveloped viruses can utilize exosome-associated proteins to form and release virions. Li et al report that HBV-envelope proteins are detected to be colocalized with AIP1/ALIX and VPS4B that are involved in the formation of exosomes, and the mutation of either of these proteins leads to blocking the release of enveloped HBV virions.33 Also, exosomes derived from IFN-α treated macrophages can transfer IFN-α-induced antiviral effect to HepG2.2.15 cells, thereby inhibiting HBV replication in hepatocytes.34,35 Exosomal miRNA-21 and miRNA-29a derived from HBV-infected HepG2-NTCP cells can suppress the expression of IL-12p35 and IL-12p40, respectively, suggesting that exosomes are essential regulators in the innate immune response against HBV infection.36 Collectively, the exosome-mediated operational mechanism provides a new understanding of the molecular basis of HBV infection.
In addition to serum-derived exosomes, this study revealed that TAF-treated MP could also produce exosomes with antiviral effects against HBV infection. It has been reported that hepatic MP accounts for nearly 70% of MP, playing a critical role in defensive response against HBV infection.37,38 In this study, we used THP-1-derived as the origin to isolate MP-derived exosomes. Consistent with the observations from Exo-serum, Exo-MP(Exo-MP (TAF) participated in TAF-induced anti-HBV response by lowering the levels of HBsAg, HBeAg, HBV DNA, and cccDNA in HepAD38 cells. TAF can be taken up intracellularly by MP and lymphocytes and then converted to active forms.39 Thus, our results demonstrated that MP is not only a “processing plant” of TAF but also an “effect amplifier” for generating exosome-based antiviral activity.
To further investigate the mechanism underlying the exosome-mediated effect of TAF, we applied RNAseq analysis to screen the potential lncRNAs related to the effect of Exo-serum. Several studies have reported that lncRNAs are involved in HBV infection.24,40,41 Relatively, rare attention is paid to the role of exosomal lncRNAs in HBV-associated processes. In this study, we identified that lncRNAs, including HOTTIP, KRASP1, and H19, were highly expressed in Exo-serum compared with Exo-serum-NC, of which HOTTIP showed the highest upregulation. Consistently, the level of lncRNA HOTTIP was also enhanced in Exo-MP(Exo-MP (TAF). Moreover, we found that lncRNA HOTTIP knockdown and overexpression reversed and enhanced the effect of Exo-MP(Exo-MP (TAF) on HepAD38 cells, respectively. In a previous study, Yılmaz Susluer et al reported that lncRNA HOTTIP is upregulated in the plasma of HBV resolved patients, suggesting that lncRNA HOTTIP might serve as a prognostic marker for HBV patients.24 Recently, a growing body of evidence has revealed the critical role of lncRNA in the response against HBV infection. Ren et al illustrate that lncRNA HOTAIR regulates HBV transcription and replication by elevating the level of transcription factor SP1.42 Feng et al demonstrated that lncRNA PCNAP1 is increased in human liver-chimeric mice and promotes HBV replication through miRNA-154/PCNA signaling.40 Moreover, lncRNA CD160 functions as an immune suppressor in CD8+ T cells by inhibiting TNF-α and IFN-γ secretion via epigenetic modulation during HBV infection.43 As such, lncRNAs are promising biomarkers for manipulating HBV infection and developing treatments coping with HBV infection.
Although this study sheds light on the role of exosomes induced by TAF treatment, several key questions remain necessary to address in future experiments. First, this study only investigated exosomes derived from CHB patients’ serum at 48 weeks after receiving daily TAF treatment. It is worthwhile to study the exosomes isolated at other time points, such as 24 weeks, 96 weeks or longer. Second, we focused on the effect of exosomes in vitro experiments. However, more valuable conclusions have to be also drawn from in vivo experiments. Thirdly, more patients samples with a broader age range should be included. Lastly, the detailed mechanisms underlying lncRNA HOTTIP need to be elucidated comprehensively in future studies.
In conclusion, the results suggest that exosome-mediated mechanisms are essential for TAF-induced antiviral activity and that exosomal lncRNA HOTTIP might be a key regulator for HBV infection. This study provides a novel understanding of the molecular mechanism underlying TAF-induced antiviral effect.
Funding
This study was supported by the Natural Science Foundation of Shandong Province, No. ZR2014HM081; Shandong Province Medical and Health Science and Technology Development Plan, No. 2015WS0378; National Natural Science Foundation of China, No. 81800444; and Natural Science Foundation of Hubei Province, No. 2018CFB415.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Dehesa-Violante M, Nuñez-Nateras R. Epidemiology of hepatitis virus B and C. Arch Med Res. 2007;38(6):606–611. doi:10.1016/j.arcmed.2007.03.001
2. Razavi-Shearer D, Gamkrelidze I, Nguyen MH, et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403.
3. Dore GJ, Cowie B. Global Hepatitis B Virus Elimination by 2030: China is Pivotal and Instructive. Oxford University Press US; 2021.
4. Suk-Fong Lok A. Hepatitis B treatment: what we know now and what remains to be researched. Hepatol Commun. 2018;3:8–19.
5. Sax PE, Wohl D, Yin MT, et al. Tenofovir alafenamide versus tenofovir disoproxil fumarate, coformulated with elvitegravir, cobicistat, and emtricitabine, for initial treatment of HIV-1 infection: two randomised, double-blind, Phase 3, non-inferiority trials. Lancet. 2015;385:2606–2615.
6. Buti M, Gane E, Seto WK, et al. Tenofovir alafenamide versus tenofovir disoproxil fumarate for the treatment of patients with HBeAg-negative chronic hepatitis B virus infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet Gastroenterol Hepatol. 2016;1:196–206.
7. Chan HL, Fung S, Seto WK, et al. Tenofovir alafenamide versus tenofovir disoproxil fumarate for the treatment of HBeAg-positive chronic hepatitis B virus infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet Gastroenterol Hepatol. 2016;1:185–195.
8. Boelens MC, Wu TJ, Nabet BY, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513.
9. Pefanis E, Wang J, Rothschild G, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789.
10. Alenquer M, Amorim MJ. Exosome biogenesis, regulation, and function in viral infection. Viruses. 2015;7(9):5066–5083. doi:10.3390/v7092862
11. Wang J, Cao D, Yang J. Exosomes in Hepatitis B virus transmission and related immune response. Tohoku J Exp Med. 2020;252(4):309–320. doi:10.1620/tjem.252.309
12. Jiao L, Qing L, Da-ming W, Xing-wang J. The association of mir-122 and mir-146a levels in serum exosomes with HBV DNA load in chronic hepatitis B patients. Chin Hepatol. 2020;25:1038.
13. Wu W, Wu D, Yan W, et al. Exosomes with miR-574 transfer anti-HBV activity mediated by the interferon from macrophage to HBV-infected hepatocyte. J Infect Dis. 2020;223:4.
14. Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15:1–14.
15. Graf J, Kretz M. From structure to function: route to understanding lncRNA mechanism. BioEssays. 2020;42:2000027.
16. Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res. 2018;7:S243.
17. Goodyear MD, Krleza-Jeric K, Lemmens T. The Declaration of Helsinki. British Medical Journal Publishing Group; 2007.
18. Li X, Zhao J, Yuan Q, Xia N. Detection of HBV covalently closed circular DNA. Viruses. 2017;9:139.
19. Rekker K, Saare M, Roost AM, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–138.
20. Garcia-Contreras M, Shah SH, Tamayo A, et al. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci Rep. 2017;7:1–10.
21. Wang G, Liu W, Zou Y, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a–MET pathway. EBioMedicine. 2019;40:432–445.
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408.
23. Durantel D. New treatments to reach functional cure: virological approaches. Best Pract Res Clin Gastroenterol. 2017;31:329–336.
24. Yılmaz Susluer S, Kayabasi C, Ozmen Yelken B, et al. Analysis of long non-coding RNA (lncRNA) expression in hepatitis B patients. Bosnian J Basic Med Sci. 2018;18:150–161.
25. Ruane PJ, DeJesus E, Berger D, et al. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of tenofovir alafenamide as 10-day monotherapy in HIV-1–positive adults. J Acquir Immune Def Syndr. 2013;63:449–455.
26. Cathcart AL, Chan HL-Y, Bhardwaj N, et al. No resistance to tenofovir alafenamide detected through 96 weeks of treatment in patients with chronic Hepatitis B infection. Antimicrob Agents Chemother. 2018;62:e01064–18.
27. Wohl D, Oka S, Clumeck N, et al. Brief report: a randomized, double-blind comparison of tenofovir alafenamide versus tenofovir disoproxil fumarate, each coformulated with elvitegravir, cobicistat, and emtricitabine for initial HIV-1 treatment: week 96 results. J Acquir Immune Def Syndr. 2016;72:58–64.
28. Agarwal K, Fung S, Seto W, et al. A phase 3 study comparing tenofovir alafenamide (TAF) to tenofovir disoproxil fumarate (TDF) in patients with HBeAg-positive, chronic hepatitis B (CHB): efficacy and safety results at week 96. J Hepatol. 2017;66:S478.
29. Ogawa E, Nomura H, Nakamuta M, et al. Tenofovir alafenamide after switching from entecavir or nucleos (t) ide combination therapy for patients with chronic hepatitis B. Liver Int. 2020;40:1578–1589.
30. Uchida Y, Nakao M, Tsuji S, et al. Significance of switching of the nucleos (t) ide analog used to treat Japanese patients with chronic hepatitis B virus infection from entecavir to tenofovir alafenamide fumarate. J Med Virol. 2020;92:329–338.
31. Lin CL, Yang HC, Kao JH. Hepatitis B virus: new therapeutic perspectives. Liver Int. 2016;36:85–92.
32. Li S, Li S, Wu S, Chen L. Exosomes modulate the viral replication and host immune responses in HBV infection. Biomed Res Int. 2019;2019:1–9.
33. Li R, Fu X, Tang Y, et al. Expression profiles of the exosomal miRNAs in the chronic hepatitis B patients with persistently normal ALT. J Centr South Univ Med Sci. 2018;43:475–480.
34. Yao Z, Qiao Y, Li X, et al. Exosomes exploit the virus entry machinery and pathway to transmit alpha interferon-induced antiviral activity. J Virol. 2018;92:e01578–18.
35. Li J, Liu K, Liu Y, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol. 2013;14:793–803.
36. Kouwaki T, Fukushima Y, Daito T, et al. Extracellular vesicles including exosomes regulate innate immune responses to hepatitis B virus infection. Front Immunol. 2016;7:335.
37. Han M, Li Y, Wu W, et al. Altered expression of interferon-stimulated genes is strongly associated with therapeutic outcomes in hepatitis B virus infection. Antiviral Res. 2017;147:75–85.
38. Wu D, Wang P, Han M, et al. Sequential combination therapy with interferon, interleukin-2 and therapeutic vaccine in entecavir-suppressed chronic hepatitis B patients: the Endeavor study. Hepatol Int. 2019;13:573–586.
39. Hermansson L, Yilmaz A, Price RW, et al. Plasma concentration of neurofilament light chain protein decreases after switching from tenofovir disoproxil fumarate to tenofovir alafenamide fumarate. PLoS One. 2019;14:e0226276.
40. Feng J, Yang G, Liu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227.
41. Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumor Biol. 2015;36:3231–3236.
42. Ren F, Ren J-H, Song C-L, et al. LncRNA HOTAIR modulates hepatitis B virus transcription and replication by enhancing SP1 transcription factor. Clin Sci. 2020;134:3007–3022.
43. Wu J, Niu Q, Yuan J, Xu X, Cao L. lncRNA‑CD160 decreases the immunity of CD8+ T cells through epigenetic mechanisms in hepatitis B virus infection. Oncol Lett. 2020;20:235–247.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.